
- Institution: Stanford Univ Med Ctr Lane Med Lib/Periodical Dept/Rm L109
- Sign In as Member / Individual
Macromolecular Therapeutics
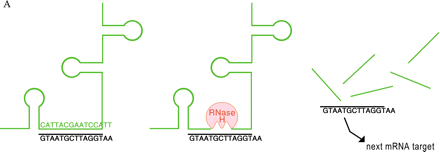
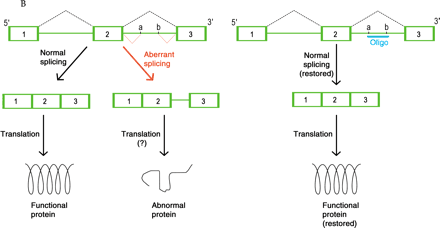
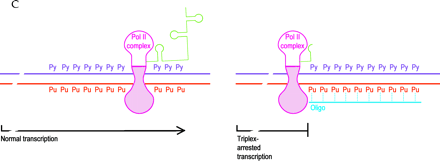
Mechanisms of oligonucleotide therapy.
A. RNase-H–mediated degradation of antisense-targeted mRNA. An antisense oligonucleotide (shown in black) that mimics a deoxyribose-phosphate backbone base pairs with a specific sequence in an unstructured region of an mRNA (green) target (left). The resulting RNA/DNA-like heteroduplex (center) is a suitable substrate for RNase H (red), which then cleaves the mRNA and opens the way to further degradation by other cellular nucleases (right). The oligonucleotide is released and cycles to other molecules of the mRNA target.
B. Inhibition of cryptic splice sites. Mutations that result in cryptic splice sites (denoted as a and b) can cause aberrant processing (indicated in red) of pre-mRNA (green) so that nonfunctional protein is produced (left). Therapeutic use of an RNA-type oligonucleotide (oligo; blue) blocks the cryptic splice site and restores normal splicing so that normal mRNA and functional protein are produced (right).
C. Triplex inhibition of transcription. The RNA polymerase (pink) complex is normally processive (left), moving along double-stranded gene sequences (purple and red) to produce mRNA (green). The presence of a triplex-forming oligonucleotide (oligo; blue) in a purine-rich tract (right; Pu, purine; Py, pyrimidine) interrupts the processivity of the polymerase complex and thus interdicts transcription (right).


