
- Institution: Stanford Univ Med Ctr Lane Med Lib/Periodical Dept/Rm L109
- Sign In as Member / Individual
Isoforms of Mammalian Adenylyl Cyclase: Multiplicities of Signaling
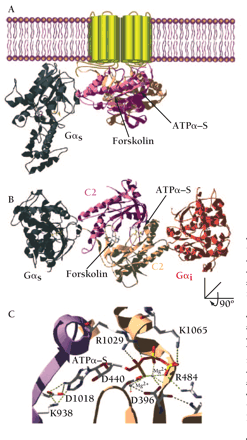
Structure of membrane-bound mammalian adenylyl cyclase bound to the activator Gαs. (A) Illustration of the crystal structure of the catalytic domain of adenylyl cyclase bound to Gαs (36) and superimposed onto the membrane-spanning region of mammalian adenylyl cyclase. Gαs•GTPγ–S in its activated form is demarcated in gray. The cyclase domains, C1 (tan) and C2 (mauve) interact and form the binding sites for forskolin and the substrate, ATP. (B) The same structure as in (A) but rotated around the x-axis of vision to give a perspective from the inner surface of the membrane. Gαi (in red) is overlaid onto the Gαs•C1•C2 structure at the pseudosymmetrically related binding site to the Gαs•GTPγ–S site. (C) The active site of adenylyl cyclase bound to the ATP analog ATPα–S(RP). Highlighted are residues that make contact with the nucleotide and that are conserved in all mammalian adenylyl cyclases. Asp396 (D396), Asp440 (D440), and Arg484 (R484) are in the C1 domain of AC5. Lys938 (K938), Asp1018 (D1018), Arg1029 (R1029), and Lys1065 (K1065) are in the C2 domain of AC2. Also indicated are two Mg2+ ions liganded by the phosphates of ATPα‴S(RP) and the two aspartate residues. The 3D structure was visualized with SwissPDBViewer™(178) and rendered with POV-RAY™ using coordinates from the Gαs•C1•C2•forskolin•ATPα–S(RP) (PDB id:1CJK) and Gαi (PDB id:1GIA) structures.


