
- Institution: Stanford Univ Med Ctr Lane Med Lib/Periodical Dept/Rm L109
- Sign In as Member / Individual
Fragment-based ligand discovery
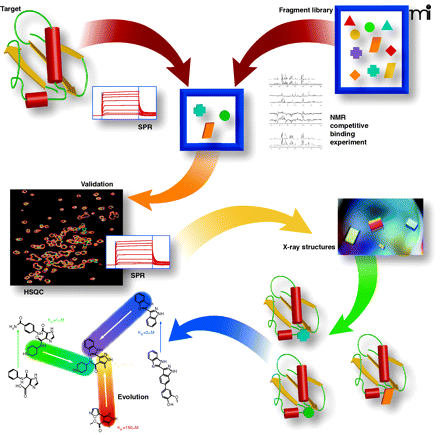
Overview of fragment-based discovery. The following description is based on processes developed and implemented at Vernalis (22, 44). A 1,200-member fragment library is screened in mixtures of up to 12 compounds for binding to the target protein by either NMR or SPR. Three NMR experiments are run (STD, CPMG, and LOGSY). A hit is defined as a fragment for which binding signals in all three experiments are reduced or abolished on addition of a competitive ligand. Alternatively, SPR is used where a robust attachment of the protein can be made to the sensor chip surface. Typically, a screen of 1,200 fragments results in between 10–100 fragment hits. Validation by HSQC and/or SPR is implemented for challenging protein–protein interaction targets, where it is necessary to use complementary biophysical measurements. Attempts are then made to determine the crystal structures of as many fragments bound to the target as possible, either by soaking apo-crystals or by co-crystallization. The resulting crystal structures define the position, binding mode, and interactions made by fragments. This information can then be integrated into structure-guided medicinal chemistry to evolve the fragments into lead compounds.


