
- Institution: Stanford Univ Med Ctr Lane Med Lib/Periodical Dept/Rm L109
- Sign In as Member / Individual
The Choline Transporter Resurfaces: New Roles for Synaptic Vesicles?
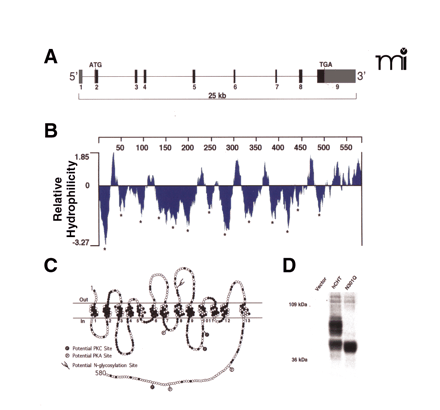
CHT revealed. (A) The hCHT gene structure comprises 9 exons encoding a 5 kb transcript distributed over a span of ~25 kb on chromosome 2. (B) Hydrophilicity analysis of the 580 amino-acid protein encoded by the 1740 bp coding region of hCHT predicts 13 transmembrane-spanning domains (TMDs, marked by asterisks). (C) The predicted transmembrane topology for hCHT includes an extracellular N-terminus and an intracellular C terminus. Amino acids conserved from humans through mice and rats to C. elegans are depicted with filled circles. There is a single predicted extracellular N-linked glycosylation site (N301) and multiple cytoplasmic protein kinase A and protein kinase C consensus phosphorylation sites [Reproduced with permission from (87) ]. (D) The predicted N-linked glycosylation site was converted to glutamine (N301Q mutant) by site directed mutagenesis and the resulting construct was transfected into COS-7 cells. Compared to the wild-type hCHT, immunoblot analysis of this N301Q mutant reveals a loss of the major glycosylated form of hCHT and an increase in the abundance of nonglycosylated CHT. Similar shifts in mobility have previously been observed following Peptide N-Glycosidase F (PNGase F) treatment of hCHT (66).


