Events
These are the meetings we are participating in and the conference we are attending, including any abstracts, posters or lectures we have presented.
October 16, 2018
State Key Laboratory of Infectious Diseases Prevention and Control —4th Academic Symposium
State Key Laboratory of Infectious Disease Prevention and Control (SKLID), Beijing, China October 16, 2018—October 17, 2018
George Garrity will be delivering a seminar on Tuesday, regarding observations on the impact of rapid changes in prokaryotic taxonomies and nomenclature. This talk highlights recent findings in taxon calling and strain classification based on NamesforLife data.
Changes in the prokaryotic taxonomy over eight months incur statistically significant changes for some test metagenomes.
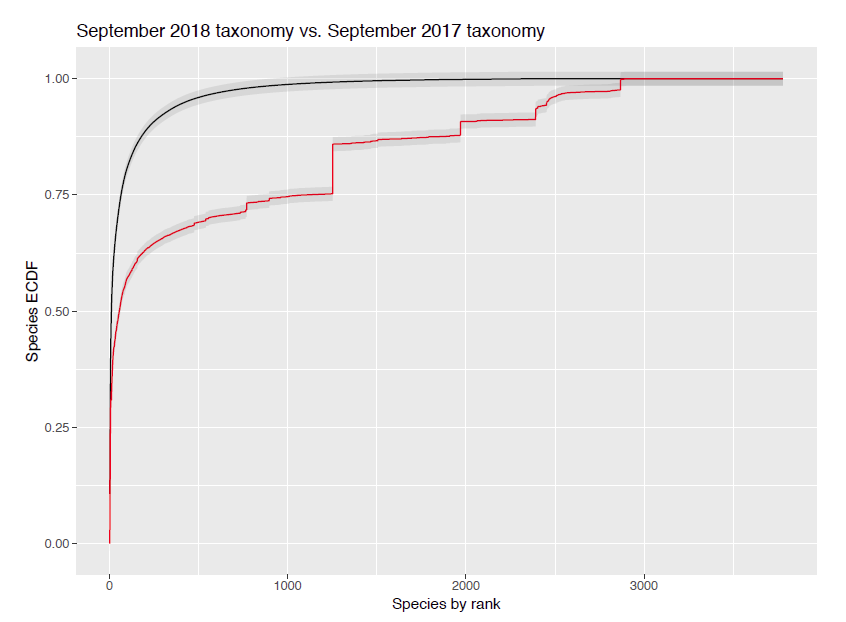
August 21, 2018
United States Culture Collections Network —2018 Meeting on Collection Data
ATCC Headquarters, Manassas, Virginia August 21, 2018—August 23, 2018
George Garrity will be delivering a presentation at 1pm on Tuesday, during the session on persistent identifiers. The topic of this talk involves making persistent connections betweens culture collections and research artifacts using the NamesforLife Information Architecture and web services.
These services provide a novel and direct means of assessing the impact of research products by individuals and research institutions that are used by the community but rarely cited. NamesforLife provides a way to correct this deficiency and objectively assess the impact of curators and resource providers.

June 28, 2018
Korean Society for Microbiology & Biotechnology —KMB 2018 45th Annual Meeting & International Symposium
Yeosu, South Korea June 27, 2018—June 29, 2018
George Garrity will be delivering the opening lecture, “Taxonomic Inference vs. Ground Truth” at this year’s KMB meeting. The lecture will be Thursday June 28th from 2:05 to 3:35pm in Rm1.
The idea of change in microbiology and other fields is nothing new. Our methods are continuously evolving, but ultimately, we need to be able to place our new findings into a frame of reference; to define our findings and to interpret the meaning of those findings.
September 10, 2017
Basel Life Innovation Forums 2017 —Innovating MedComms
Congress Center, Basel, Switzerland September 10, 2017—September 13, 2017
George Garrity will be presenting on two topics during the Innovating MedComms panel: How to ensure content quality in a world of overwhelming scientific complexity, 1:30pm-2:30pm (Machine learning-based tools for peer review) and Scientific discovery In the Machine Age: New tools for competitive advantage, 3:30pm-4:30pm (Machine learning tools for discovering scientific content). Both sessions are in the Shanghai 1 room, and videos will be made available after the event.
The first session (Machine learning tools for discovering scientific content) will showcase how novel semantic tagging and document classification methods can be used to enrich content by unobtrusively integrating externally curated resources and references. Further discussion will explore how these curated resources can serve as hidden metrics that provide a supplementary measure regarding the significance of various research artifacts or concepts in a given field of study.
The following session focuses on applying machine learning tools to the peer review process.
George Garrity reasons that most people underestimate the amount of work that goes into the process. “The publisher distributes your content, they polish it, they make sure there’s an archival version, but they also provide all the necessary quality control, and this is typically done by peer review,” he said.
The peer review process is essential for checking that valid arguments and conclusions are present, with appropriate priority, provenance and originality. However, it can be costly and very time-consuming, thus there is great interest in automating as much of the process as possible.
Hoping to do just that, a suite of tools from NamesforLife allows processing of a raw manuscript in mere minutes, validating facts, structure, terminology and cited resources, and annotating any “red flags”. The automation can then extend to the peer review stage, cross-checking the intended submission with a pool of some 40,000 documents in order to identify candidate reviewers based on relevant publication records.
The process removes selection bias, screens for conflicts of interest, and tracks ongoing reviewer performance. What’s more, it keeps up-to-date contact information for reviewers, and constructs a compelling email to send to the reviewer to encourage their participation.
June 28, 2017
Korean Society for Microbiology & Biotechnology —KMB 2017 44th Annual Meeting & International Symposium
BEXCO, Busan, South Korea June 28, 2017—June 30, 2017
George Garrity will be delivering the opening lecture, “Some Thoughts and Observations on ‘Taxon Calling’” at this year’s KMB meeting. The lecture will be Wednesday June 28th from 12:30 to 1:10pm at APEC Hall.
The focus of this lecture will be to demonstrate the value of a well-curated and carefully annotated reference database that can be used to evaluate existing and new methods of identifying and assigning names to prokaryotic taxa which can serve as a standard and be used for routine re-annotate and updating of existing metagenomes and microbiomes at a much finer grain of resolution that is currently used.
May 31, 2017
Society for Scholarly Publishing 39th Annual Meeting —Striking a Balance: Embracing Change While Preserving Tradition in Scholarly Communications
Westin Boston Waterfront, Boston, Massachusetts May 31, 2017—June 2, 2017
NamesforLife has a booth at the SSP 2017 annual meeting this year. Stop by booth number TT7 for a demonstration of how our tools are being used by early adopters, how our approach might meet your needs for semantic enrichment of your content, and how you can help us shape forthcoming features.
Our software architect, Charles Parker, and our founder, George Garrity will be available every day of the conference for questions and product demonstrations.
Online tools have improved the efficiency of many parts of the editorial workflow, but also place pressure on publishers to perform new tasks in the service of authors and readers. These include identifying suitable editors and peer-reviewers and ensuring technical accuracy of published content. These tasks require a high level of domain knowledge that is often in short supply. We offer services to fill these gaps that can be integrated into existing editorial platforms.
March 8, 2017
London Book Fair 2017 —Advancing Editorial Productivity with NamesforLife Production Workflow Solutions
Olympia, London, England, United Kingdom March 14, 2017—March 16, 2017
NamesforLife has a booth at the London Book Fair this year. Please stop by Stand 3B36 for product demonstrations and join George Garrity at the Tech Theater on Tuesday March 14th at 12:15pm for a seminar on how our tools are being used by early adopters to improve editorial efficiency. The presentation will be posted here after the seminar.
NamesforLife semantic services provide scientific and technical publishers with standards-based editorial workflow solutions that enhance the value of content to readers while reducing the efforts of authors, peer-reviewers and editors to produce technically accurate content.
Our semantic annotation services save time at each stage of the editorial process and continue to add value after publication. Detection and correction of errors at the earliest possible stage of content production results in significant improvement of document throughput and substantial cost savings.

November 29, 2016
Defense Innovation Summit 2016 —Autonomous Systems
Austin, Texas November 29, 2016—December 1, 2016
Charles Parker and George Garrity will be attending the Defense Innovation Summit this year. We will be presenting an overview of our recent work on poster 313, “Knowledge Extraction from Mixed-Precision Information”, during Poster Session I Tuesday afternoon from 2:30pm-3:15pm. We are actively seeking commercial partners to bring this technology to market.
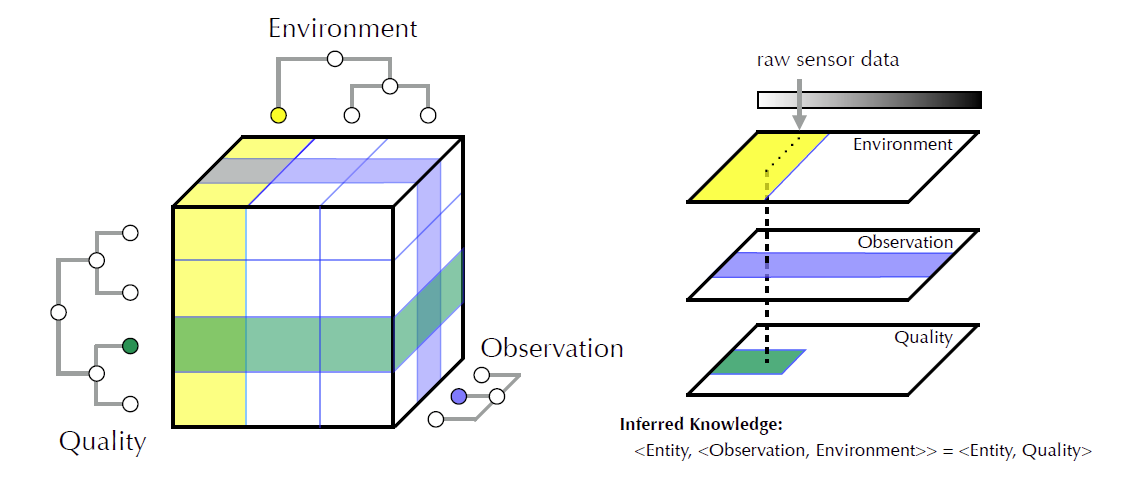
A fundamental barrier to effective human-machine communication is the lack of a shared, unambiguous language that is understandable to humans and precise enough for machine reasoning. The knowledge of domain experts is aggregated from a variety of information sources, ranging from raw text or data to structured and normalized databases (Mixed Precision Information; MPI).
We introduce a novel standards-based method for extracting knowledge from MPI to provide knowledge workers and machine reasoners with verifiable interpretations of observational data.
Our approach combines semantic and semiotic methods to represent information at multiple levels in concept hierarchies, “slice” and aggregate concepts to represent information consistently for ambiguous human language and reasoners, provide multiple entry points for information (term, concept, data), provide attachment points for reasoning over rules and axioms and accommodate multiple interpretations of information.
April 12, 2016
London Book Fair 2016
Olympia, London, England, United Kingdom April 12, 2016—April 14, 2016
NamesforLife will be attending the London Book Fair this year. Although we are not presenting this year, we have demonstrations available for our upcoming reviewer services.
March 1, 2016
Genomic Science Program (GSP) 2016 —Contractors-Grantees Meeting XIV
Tysons, Virginia March 6, 2016—March 9, 2016
Charles Parker and George Garrity will be presenting poster 147 (“Semantic Index of Phenotypic and Genotypic Data”, Abstract Book, pages 256-257) highlighting their team’s recent work during the Monday evening mixer (5:00pm-7:00pm) in Tyson’s Ballroom.
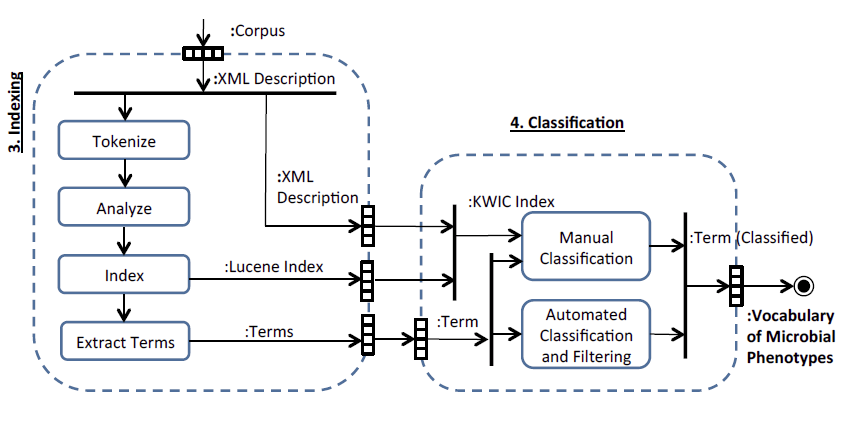
During the course of this project we developed many software components that overcome specific technical barriers in terminology management, text mining, information extraction, knowledge transformation, entity recognition, document classification and annotation. The individual tools (N4L::Guide, N4L::Scribe, the Taxonomic Abstracts, Taxomatic, the KWIC Index and the Semantic Desktop) were implemented using W3C standards and recommendations (SPARQL, RDFS, RDF, OWL2, SKOS, SKOS-XL, XML, XSL, XSD, SPIN, OWL RL, DOI/CrossRef, CORS) and commercially-compatible FOS frameworks (Java, Apache, PostgreSQL, Virtuoso OSE, Jena/ARQ, SPIN Reasoner). We are integrating these components into a single software suite that can support a variety of document analysis needs.
Backed by the Fairview Research Alexandria platform (CLAIMS Global Patent Database), this analysis suite has access to the full text of the worldwide patent literature. We have demonstrated the ability to reverse-engineer the diagnostic phrases that human indexers use to classify large corpora of technical documents, and to measure both the quality of previously annotated documents and the cohesion of individual document classifications. Our software provides a novel way to navigate and bridge multiple classification systems.
Our continued collaborations with the Joint Genome Institute, Fairview Research/IFI Claims and Oak Ridge National Laboratories provide excellent opportunities to test and refine the capabilities of this analysis suite while raising the visibility of other federal funded projects by completing the semantic linking between projects, entities and publications.
May 1, 2015
17th Workshop of the Genomic Standards Consortium (GSC17) —Standards for the Microbial Dark Matter (uncultured microbial life)
Department of Energy Joint Genome Institute, Walnut Creek, California May 4, 2015—May 6, 2015
Charles Parker will be presenting a poster at the GSC Workshop on May 5th.
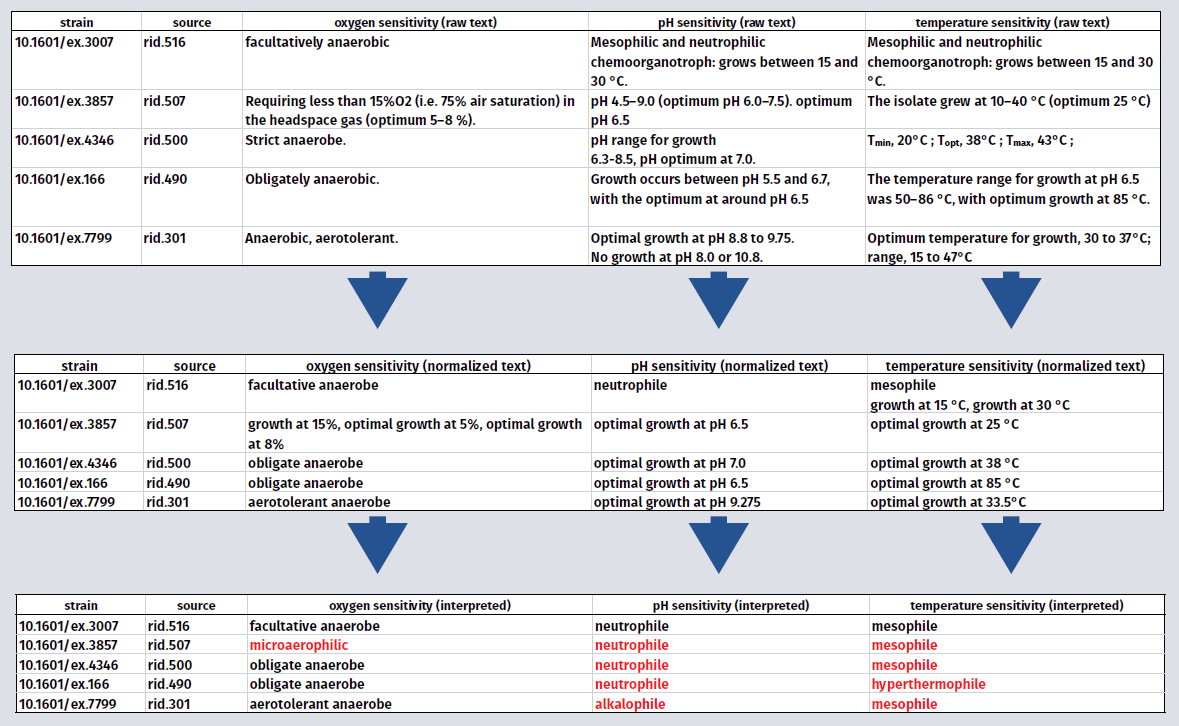
Despite significant improvements in genome annotation, many assertions are hypothetical and may lack experimental support. The taxonomic literature for prokaryotes contains a wealth of experimental phenotypic data, but that knowledge is currently in a form that does not lend itself to integration with databases or ontologies.
Our knowledge base is designed to address these problems by providing reference phenotypic data for nearly all type strains of Bacteria and Archaea, based on concepts and observational data drawn from the primary taxonomic literature (the corpus of literature that supports our up-to-date taxonomy and strain database).
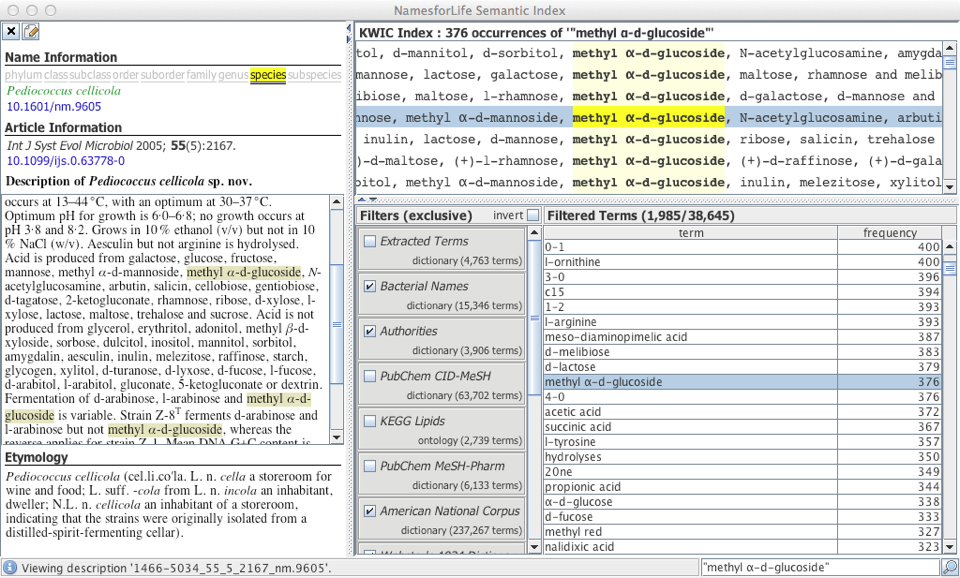
We developed software (Semantic Desktop) to extract putative feature domain vocabularies from this corpus, we have since developed this vocabulary into a precise thesaurus of phenotypic terms, which will ultimately conform to W3C SKOS-XL semantics, providing a link between the language of microbial phenotype, the semantic web and existing NamesforLife services.
April 30, 2015
Patent Users Information Group Annual Conference and USPTO-EPC CPC Annual Meeting —From Search Strategy to Business Strategy: Domestic and International Practices, Styles, and Viewpoints
Westin Lombard Yorktown Center, Lombard, Illinois May 1, 2015—May 7, 2015
NamesforLife is attending the USPTO-EPO CPC Annual Meeting with Industry Users on May 1st, as well as the PIUG Annual Conference from May 2-7.
Our company has developed several innovative software components to overcome technical barriers in text mining, information extraction, document classification and annotation.
Our technology arose from a need to support thesaurus construction, vocabulary integration and ontology development. As a result, we have created bridges between document analytics and important industry standards for knowledge representation. Our patented technology produces high-quality data sets from scientific, medical and legal literature via its partnerships with the academic publishers, and Fairview Research, LLC.
Our classification tools provide novel ways to navigate and bridge various patent classification systems, enabling more precise classification and integration with additional proprietary classifications.
These individual software components have been integrated into a single platform that can support a variety of document analysis needs. Our software may be deployed in a web service container, as a desktop application, or extended/integrated with third party software via our developer API.
Backed by the Fairview Research Alexandria platform (CLAIMS Global Patent Database), this analysis suite has access to the full text of the worldwide patent literature.
February 20, 2015
Genomic Sciences Program (GSP) 2015 —Contractors-Grantees Meeting XIII
Tysons, Virginia February 22, 2015—February 25, 2015
Charles Parker and George Garrity will be presenting poster 222 (“Semantic Index of Phenotypic and Genotypic Data”, Abstract Book, page 333) during Tuesday evening’s mixer (5:00pm-7:00pm) in Tyson’s Ballroom. We will be highlighting our team’s recent work on Knowledge Extraction from scientific literature.
Our core technical objectives are to: (1) build a database of normalized phenotypic descriptions using the primary taxonomic literature of bacterial and archaeal type strains, (2) construct an ontology capable of making accurate phenotypic and environmental inferences based on that data, and (3) improve the visibility and accessibility of publicly-available research data.
This project is tightly coupled with ongoing DOE projects (the Genomic Encyclopedia of Bacteria and Archaea, the Microbial Earth Project, the Community Science Program) and with two key publications, Standards in Genomic Sciences (SIGS) and the International Journal of Systematic and Evolutionary Microbiology (IJSEM).
The scope of this project covers many technical fields, including text-mining, Information Extraction, Natural Language Processing, indexing & search, terminology & ontology development, machine reasoning, semantic analysis, sequence analysis and taxonomic classification.
September 6, 2014
International Union of Microbiological Societies Conference 2014 —International Congress of Bacteriology and Applied Microbiology
Convention centre (Palais des congrès), Montréal, Québec, Canada July 27, 2014—August 1, 2014
George Garrity and Charles Parker will be attending the International Congress of Bacteriology and Applied Microbiology at the IUMS 2014 conference. We will be submitting a draft of the next edition of the International Code of Nomenclature of Prokaryotes.

May 19, 2014
Second Workshop of the United States Culture Collections Network —Fusarium Research Laboratory, Penn State University
State College, Pennsylvania, United States May 19, 2014—May 21, 2014
George Garrity presents “Standards to Promote Data Interchange in the Life Sciences”.
This discussion will focus on emerging data, metadata, publishing and web standards and explore how collections might adopt these standards as part of their strategy in developing and delivering interoperable information products to the market.
...these issues are ultimately dependent upon accurate and properly curated reference material, further discussion included the use of standards in managing collection materials. Different standards were described including self imposed standards such as nomenclature and also external standards for reference material, process optimization, and data management.

January 16, 2014
Mathematical, Statistical and Computational Aspects of the New Science of Metagenomics —Isaac Newton Institute for Mathematical Sciences, University of Cambridge
Cambridge, England, United Kingdom March 24, 2014—March 28, 2014
George Garrity presents “Reasonable names and reasonable terms for Bacteria and Archaea”.
This presentation will focus on the development of a generalized semantic model that has been developed to disambiguate biological nomenclature and to provide both humans and machines with direct access to the correct information about all of the validly named prokaryotic taxa. Current research efforts on developing an ontology of microbial phenotypes, which supports machine reasoning, will also be discussed.
January 15, 2014
Genomic Sciences Program (GSP) 2014 —Contractors-Grantees Meeting XII
Arlington, Virginia February 9, 2014—February 12, 2014
Charles Parker and George Garrity will be presenting poster 170 (“Semantic Index of Phenotypic and Genotypic Data”, Abstract Book, pages 297-298) during Tuesday evening’s mixer (5:00pm-7:00pm) in Independence Center. We will be highlighting our team’s recent research on Information Extraction (IE), reasoning and ontology query.
This project has presented technical challenges that require creative solutions across several areas of information science.
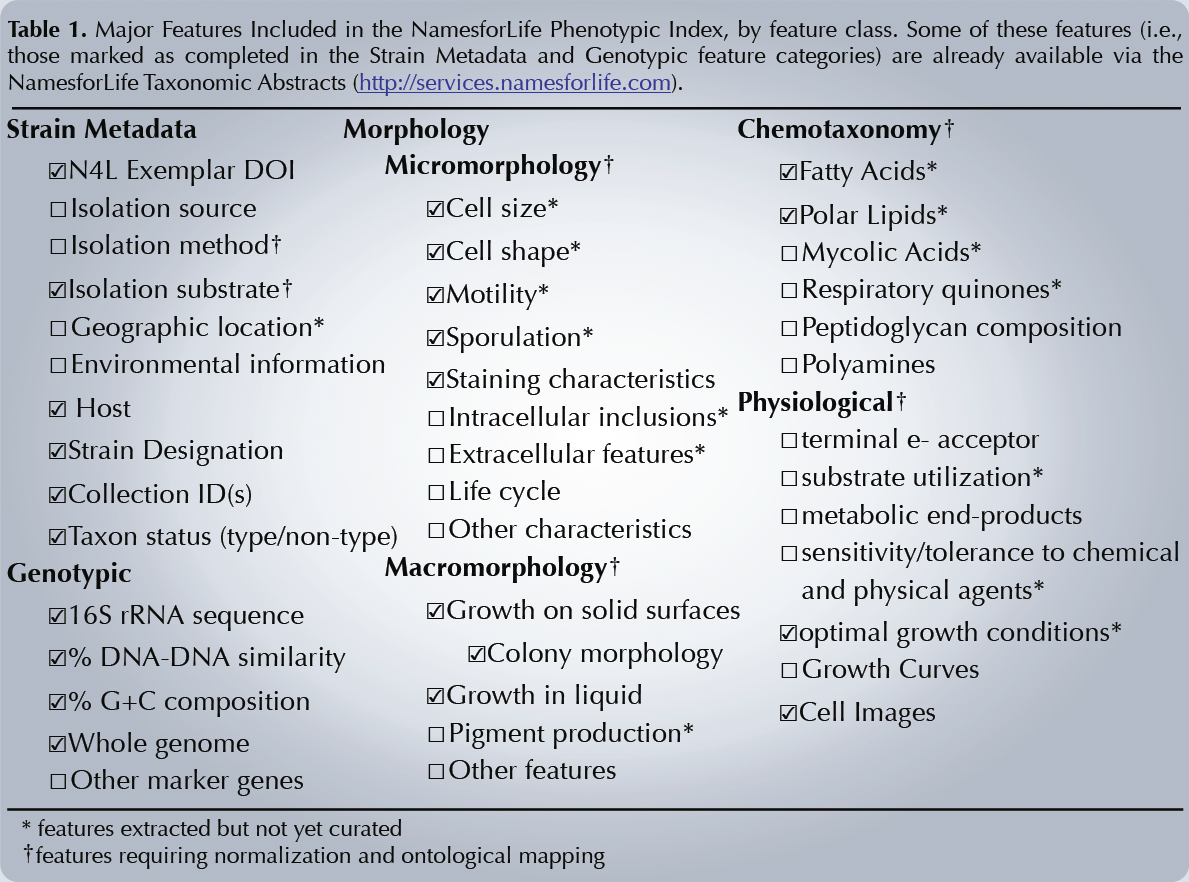
Many ontologies consist of a large thesaurus of terms in a narrowly-defined domain and do not contain any reasoning capability beyond the taxonomic structure of the vocabulary and relations among concepts. Our objective is to develop an ontology that covers many broad feature domains and contains axioms encoded in first order logic that enable reasoning and inference over sparse phenotypic data, even in feature domains that contain partially-overlapping concepts and terms that map to undefined ranges of environmental conditions. In order to accomplish this, we have developed a core ontology model that maps between imprecise phenotypic features and precise environmental data.
In our current work, we are applying these novel modeling techniques to encode Tbox axioms for automatically resolving ambiguity attributed to the semantic equivalence and imprecision of phenotypic terms arising in literature. These axioms will enable reasoners to make appropriate inferences over the ontology and phenotypic data. We are also developing a query and retrieval service linked to the ontology that will provide researchers with consistent, accurate interpretations of these data that are usable for predictive modeling and in other research and commercial applications.
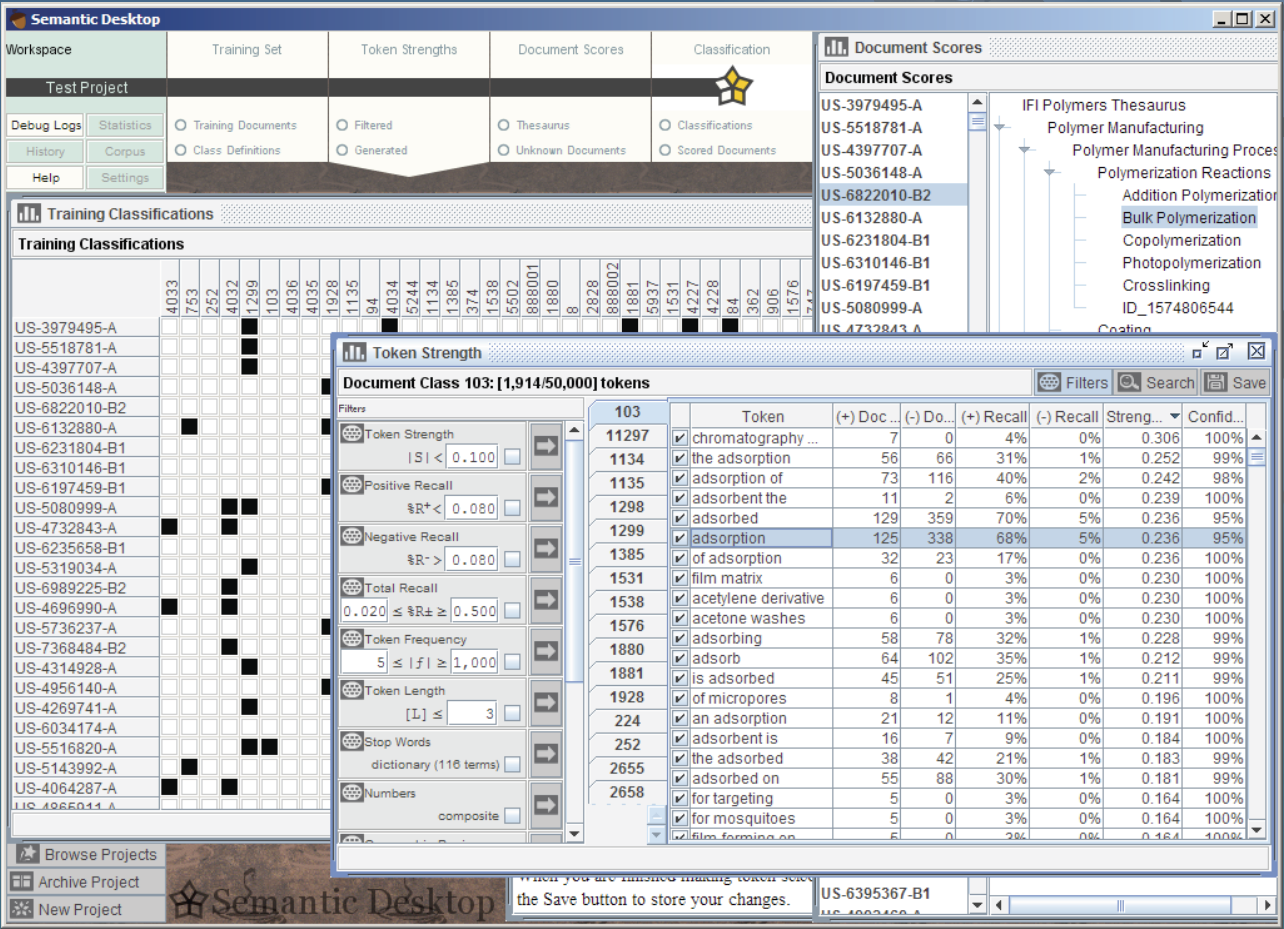
Several additional software components were developed to overcome technical barriers that arose during this project. Originally implemented as command-line utilities for vocabulary extraction, annotation and document analysis, we are now developing these into a commercial semantic desktop application for document/corpus analysis and for bootstrapping terminology/ontology development.
October 15, 2013
Society for Industrial Microbiology and Biotechnology —RAFT X: Recent Advances in Fermentation Technology
Marco Island, Florida November 3, 2013—November 6, 2013
George Garrity and Charles Parker will be presenting posters (“Global commercialization trends of microbial products and processes” and “A semantic index of phenotypic and genotypic data”) at the RAFT X conference. The poster session will be in the Capri Ballroom from 5:00pm-7:30pm Monday evening. The posters are also available to attendees on the RAFT-X meeting site.
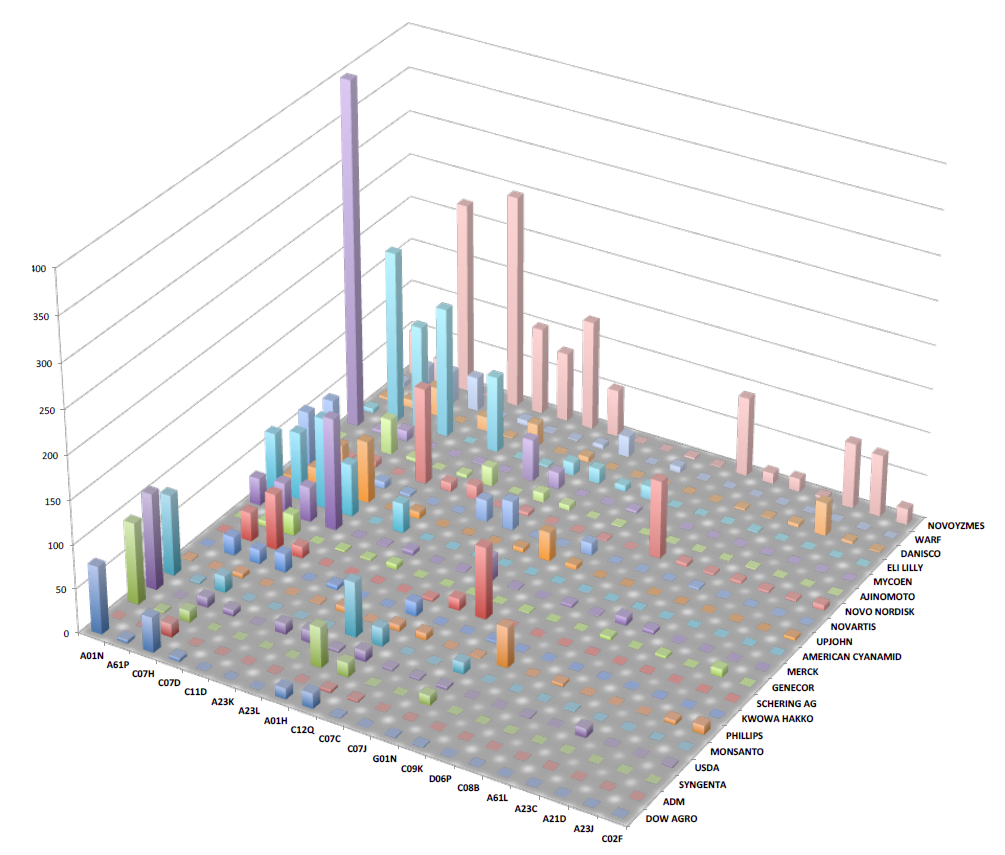
Our objective is to make the connections between strains and the patent literature easy to navigate and to make the information about patented microbial products and processes more readily discoverable. We recently completed a first pass through the USDA ARS Patent Collection (NRRL Collection, Peoria, IL). Using proprietary text mining methods, we were able to identify global commercialization trends in 162 technology classes over a 70 year time span by following more than 4,000 distinct NRRL strains referenced by over 16,000 US and foreign patents drawn from a corpus of over 80 million patent documents.
October 7, 2013
BioCreative IV Challenge and Workshop —BioCreative: Critical Assessment of Information Extraction in Biology
Bethesda, Maryland October 7, 2013—October 9, 2013
George Garrity presented an overview of the text mining approaches employed by NamesforLife during the DOE Panel on October 8th, 2013.
How might one maintain quality, consistency and usability of stored observational data over time, knowing that both the information and the underlying data are fluid and often inconsistent or even contradictory?
While text mining, natural language processing and machine reasoning are all thought of as computational problems, our experience teaches that the human element, provided by Subject Matter Experts and data curators is crucial if one is to obtain useable and meaningful results. Subject Language Terminologies (SLTs) are dynamic and may contain terms that have many nuanced meanings.
We have developed a generalized process to mitigate these challenges that includes a flexible data model, document analysis methods, and a workflow.
March 24, 2013
Intellectual Property Rights Workshop —University of Arizona
Tucson, Arizona April 26, 2013—April 26, 2013
George Garrity presents an overview of NamesforLife technology, services and data products.
NamesforLife provides web services for editorial offices. These services are designed to have minimal impact on production workflows, by providing multiple access points that can be integrated at any point in a content production workflow.
We also offer consulting services in terminology and taxonomy development, including management of Subject Language Terminology, QA/QC, data cleaning, linking and annotation, and ab-initio development of vocabularies.
We have several professionally curated data products available for licensing, as well as a patented method for serving terms, names and associated information over unique identifiers.
February 24, 2013
Genomic Sciences Program (GSP) 2013 —Contractors-Grantees Meeting XI
Bethesda, Maryland February 24, 2013—February 27, 2013
Charles Parker and George Garrity will be presenting a poster (“The NamesforLife Semantic Index of Phenotypic and Genotypic Data”) during the evening mixers (5:00pm-7:00pm) on Monday and Tuesday. We will be highlighting our team’s recent research on Information Extraction (IE) and automated thesaurus construction.
Please note that due to federal travel restrictions, this meeting’s attendance and scope will be limited, and no abstracts document will be published. We appreciate the folks from Oak Ridge National Labs, who took a bus all the way from Tennessee to attend this meeting!
Phenotypic data needs to be viewed from an historical perspective to understand not only what was measured but how it was measured (growth on substrate vs. hydrolysis of indicator compound). It is also important to know which methods were applied and whether different methods within an array of data are measuring the same trait, and if so, whether the results are comparable.
The Phenotypic Index will address these issues by tying together observations under specific sets of growth conditions, supporting faceted search, retrieval and comparison of differentiating characteristics between (and within) taxonomic groups. Each phenotypic observation will be linked to a strain via a NamesforLife Exemplar DOI (Digital Object Identifier), which is directly linked to an actively maintained taxonomy and nomenclature.
January 23, 2013
NamesforLife Phenotypic Ontology —Argonne National Laboratory
Chicago, Illinois January 23, 2013—January 23, 2013
Dr. George Garrity presents NamesforLife’s progress toward a phenotypic ontology for Bacteria and Archaea.
December 31, 2012
DOI mediated semantic services —Scientific, Technical and Medical Publishers New Technologies Meeting
London, England, United Kingdom December 1, 2012—December 1, 2012
George Garrity will be presenting a five-minute overview of the NamesforLife publisher services at the 2012 STM conference.
Our goal is to provide on-demand access to information so your authors, reviewers, readers and editors can read like a Subject Matter Expert.
December 30, 2012
A potential semantic service layer for DOI RAs —International DOI Foundation Board Meeting
Oxford, England, United Kingdom December 1, 2012—December 1, 2012
George Garrity will be presenting the NamesforLife semantic annotation services at the 2012 IDF board meeting.
At the core of our services is a proprietary data model using DOIs to deliver semantic services into a publisher’s content, either through embedded links or transient links that are created on-the-fly. This allows us to apply independently managed terminologies to a digital library immediately and to provide real-time content enhancement rather than a posteriori annotation of a body of literature.
December 29, 2012
Phenotypic Dark Matter —Danish Technical University
Lyngby, Denmark December 1, 2012—December 1, 2012
December 28, 2012
Phenotypic Dark Matter —Deutsche Sammlung fur Mikroorganismen und Zellkulturen
Braunschweig, Germany December 1, 2012—December 1, 2012
December 27, 2012
DOIs, Kbase and NamesforLife —Webinar
Germantown, Maryland December 1, 2012—December 1, 2012
February 24, 2012
Genomic Sciences Program (GSP) 2012 —Contractors-Grantees Meeting X
Bethesda, Maryland February 26, 2012—February 29, 2012
Charles Parker and George Garrity will be presenting poster 228 (“The NamesforLife Semantic Index of Phenotypic and Genotypic Data”, Abstracts Book, pages 183-184) during the Monday evening mixer (5:30pm-8:00pm) in the Grand Ballroom. We will be highlighting our team’s recent research on text mining and automated vocabulary extraction.
The long-term objective of this STTR project is to develop a semantic index of bacterial and archaeal phenotypes that can be used to augment annotation efforts and to provide a basis for predictive modeling of microbial phenotype. The index is based on published descriptions of taxonomic type and non-type strains that have been the subject of ongoing genome sequencing efforts as this will provide a mechanism whereby hypotheses can be tested and reproducibility verified. This project is tightly coupled with ongoing DOE projects (Genomic Encyclopedia of Bacteria and Archaea, the Microbial Earth Project, the Community Sequencing Project) and with two key publications, Standards in Genomic Sciences and the International Journal of Systematic and Evolutionary Microbiology. The first step towards accomplishing this goal, and the primary objective of this Phase I project is the development of a draft vocabulary.
November 7, 2011
eXtyles User Group Meeting
Boston, Massachusetts November 11, 2011—November 11, 2011
Dr. George Garrity will be presenting a case study of NamesforLife at the 2011 XUG Meeting.
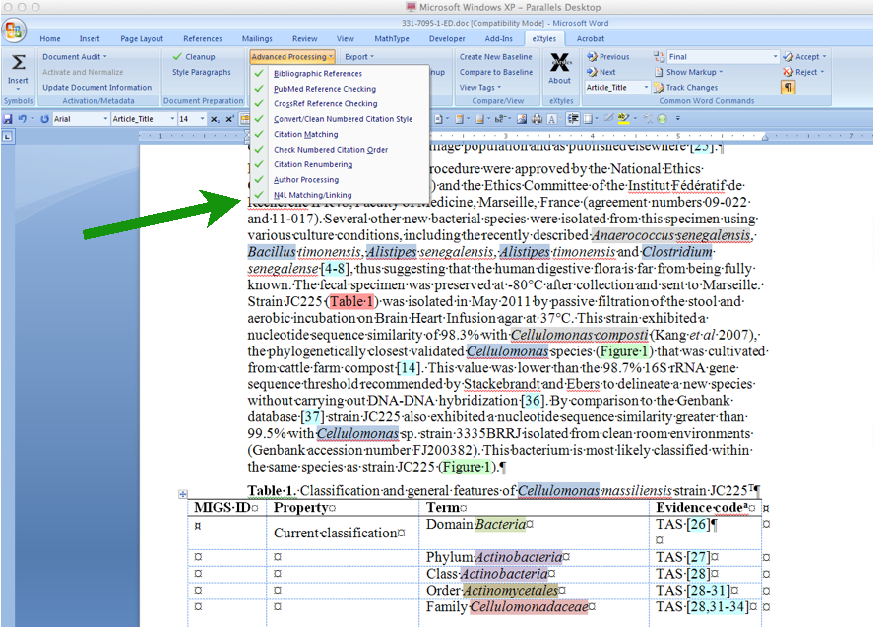
This case study will discuss integration of NamesforLife’s DOI-based semantic resolution services with eXtyles. The NamesforLife tool is designed to provide editors and authors with direct access to expertly maintained information about biological names and other dynamic terminologies as a part of the editorial process, to automatically resolve any instances of ambiguity, and to embed DOIs directly into XML instances so that readers have direct access to rich contextual information associated with each name, without having to leave the article they are reading.

October 17, 2011
SyMBIOTA: Synergy in Microbiota Research —Workshop II: Methods to Study the Human Microbiome
University of Toronto, Ontario, Canada October 17, 2011—October 18, 2011
Dr. George M. Garrity will be presenting the keynote lecture, “Distorted Realities”, during the Bioinformatics session on Monday at 9:15am.
September 6, 2011
IUMS Bacteriology and Applied Microbiology Congress —The Unlimited World of Microbes
Sapporo, Japan September 6, 2011—September 10, 2011
Dr. George M. Garrity will be presenting Plenary Lecture 4 for this conference on September 7th.
May 1, 2011
PIUG 2011 Annual Conference —Best Practices Beyond Free-text: The Value of Indexing and Classification when Searching and Analyzing Patents
Cincinnati, Ohio May 21, 2011—May 26, 2011
George M. Garrity will be presenting a lecture on applying NamesforLife semiotic analysis to Fairview’s Alexandria database during the Tuesday morning session (Indexing Patent Literature Using Semiotic Fingerprints).
April 1, 2011
Genomic Sciences Program (GSP) 2011 —Contractors-Grantees Meeting IX
Crystal City, Virginia April 10, 2011—April 13, 2011
Charles Parker and George Garrity will be presenting poster 117 (“Semantic Indexing of the Green Technology Patent Literature”, Abstracts Book, page 90) during the Tuesday evening mixer (5:30pm-8:00pm) on the Independence Level (Independence Center B). We will be highlighting our team’s recent research on semiotic document classification.
As DOE research on biofuels, bioremediation and carbon sequestration moves from the laboratory into production or commercial environments, a number of important policy and business decisions must be made that demand correct information.
An awareness of developments in the field requires a thorough review of both bodies of literature. NamesforLife is building tools to simplify such searches, using its proven approach to indexing through the creation of persistent links to externally managed terminologies that common to both bodies of literature. This approach integrates well with existing commercial, academic and USPTO data mining capabilities.
February 1, 2011
BioSystematics 2011
Berlin, Germany February 21, 2011—February 27, 2011
Charles Parker will be presenting a poster and demonstrating the NamesforLife services at the software bazaar on Thursday from 10:30am-3:00pm in the Yale-Princeton room. George Garrity will be present a 20 minute talk on Standards in Genomic Sciences on Friday evening from 6:00pm-6:20pm in the Princeton room.
Our semantic tagging web service, N4L Scribe, is now available. It tags bacterial names in any well-formed XML document with forward-linking Digital Object Identifiers. The service sits at the core of the server-side content enablement for N4L Guide, and is intended for integration into existing publication workflows. Plug-ins are currently in development for several ubiquitous word processing and desktop publishing applications as well. The service can be tested out for free on our web site with a NamesforLife account.
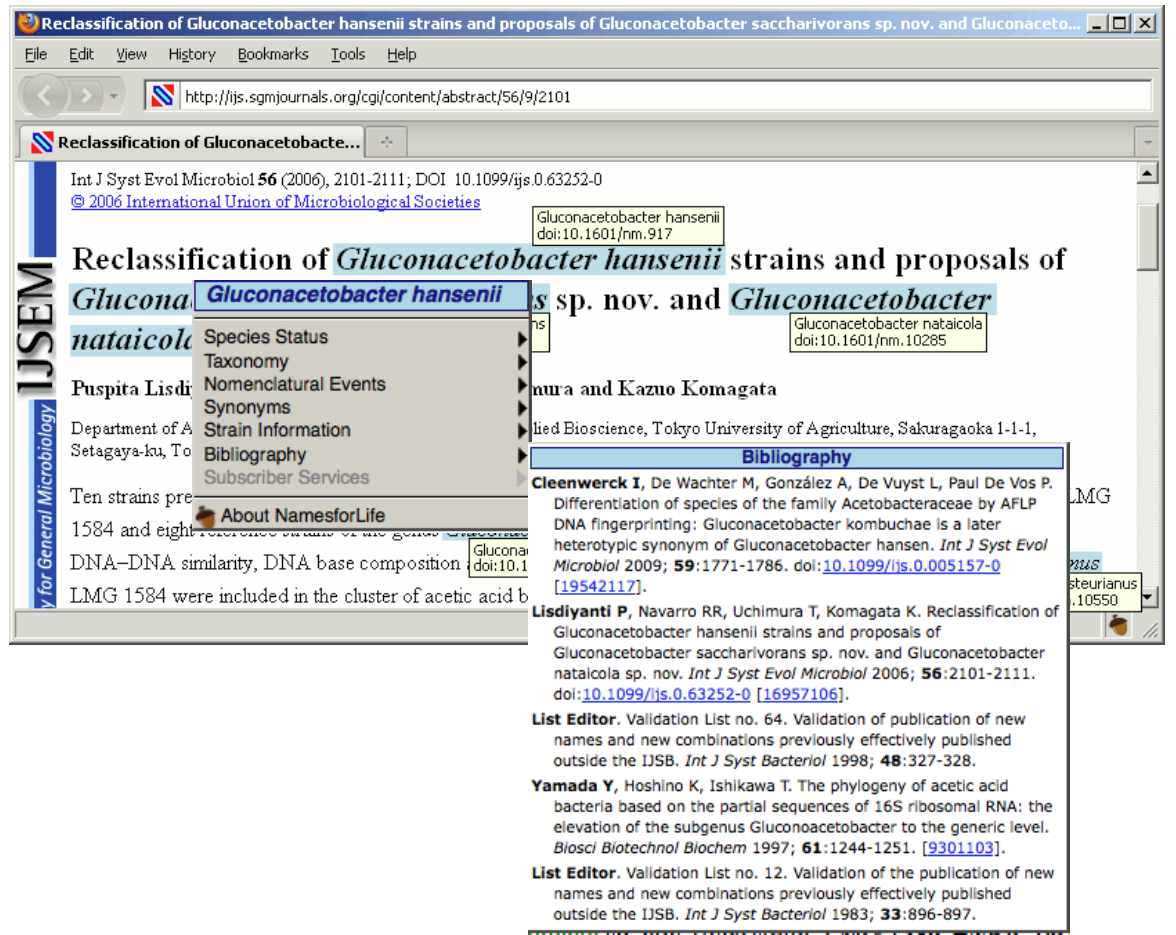
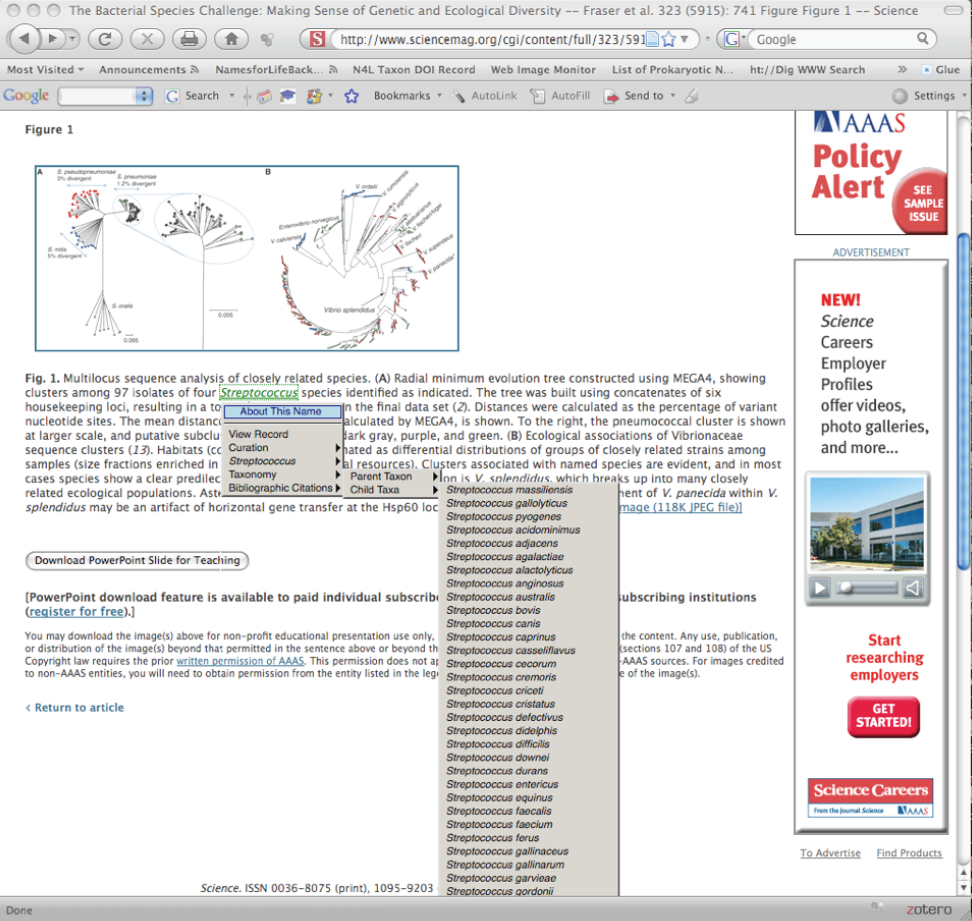
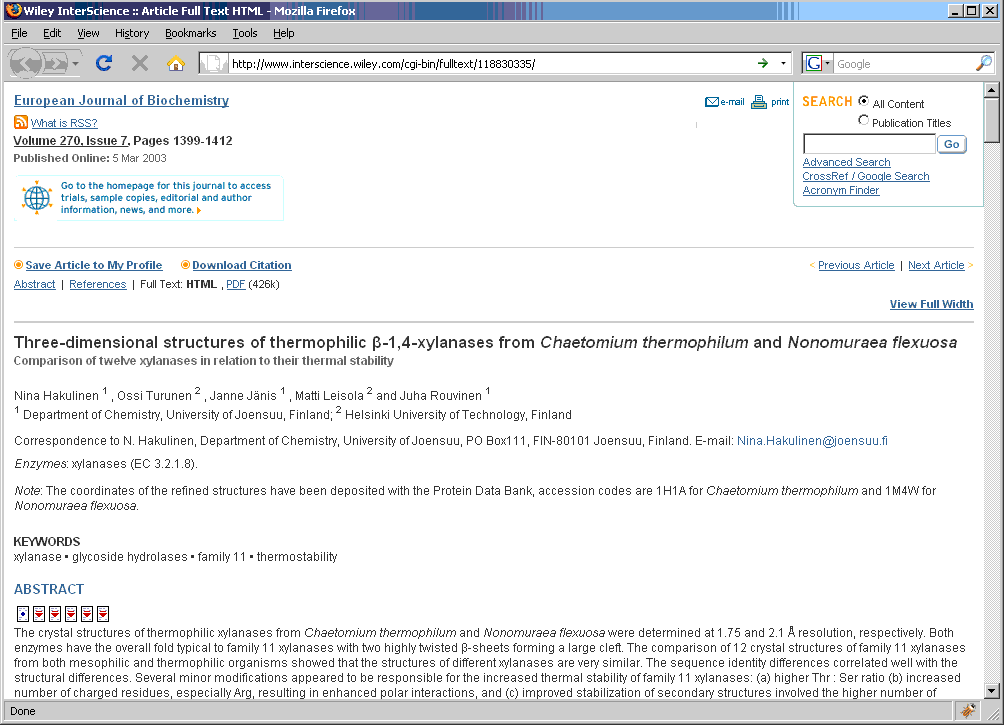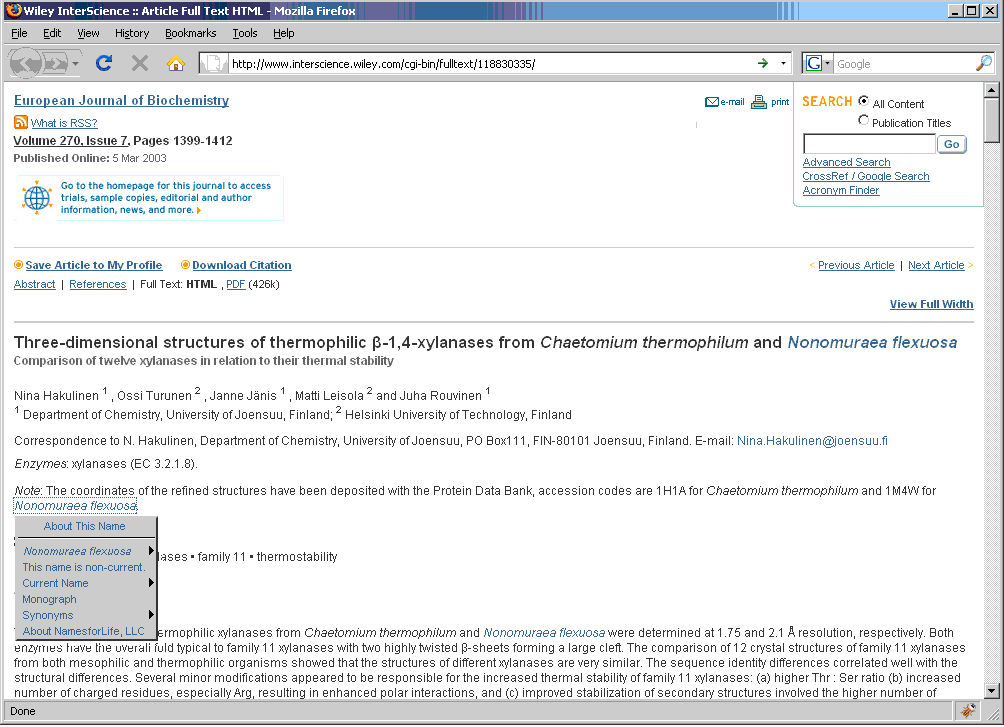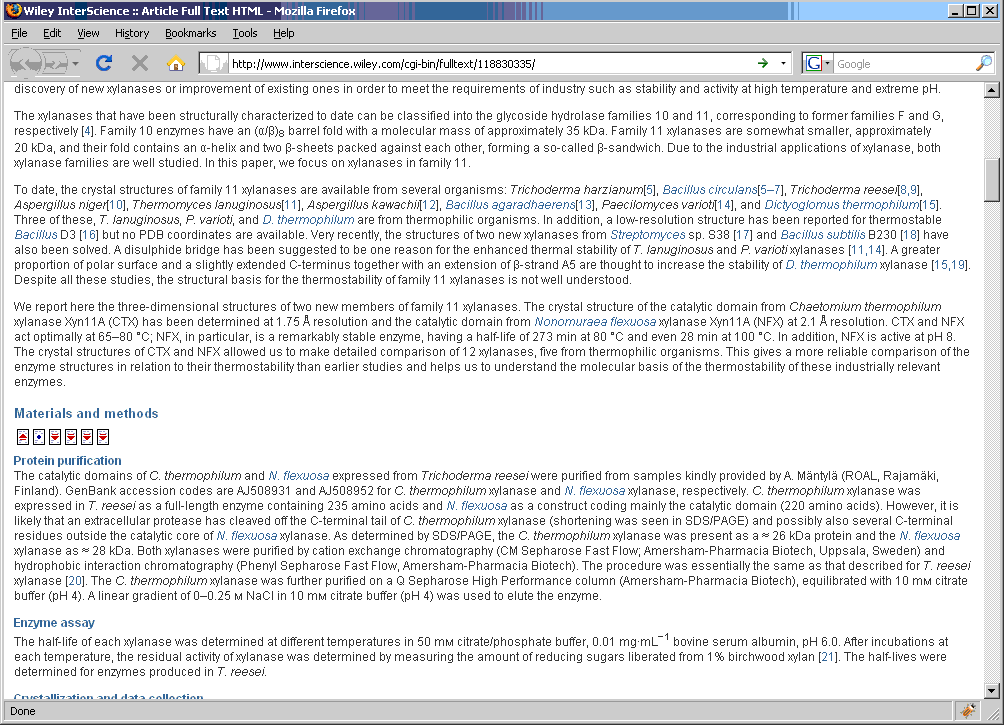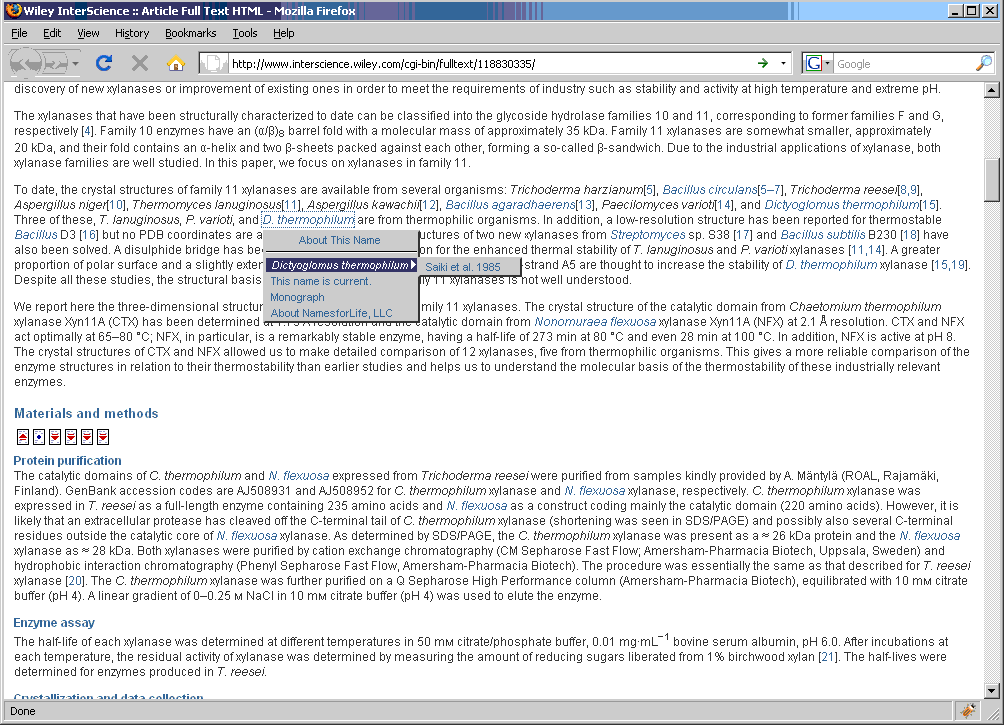
The N4L Guide browser add-on detects and links bacterial names to the N4L database, providing up-to-date nomenclature, strain and genome information, and a full bibliography. The screenshots below demonstrate the use of this tool on an IJSEM article. Instructions for installing and using this tool can be found at the NamesforLife website.
October 1, 2010
PIUG 2010 Northeast Conference
Hyatt Regency, New Brunswick, New Jersey October 11, 2010—October 15, 2010
Charles Parker from NamesforLife will be attending the main meeting and exhibition for the Patent Users Information Group Northeast conference on Tuesday, October 12th. The PIUG Northeast Conference brings together experts in the area of chemistry/biology, non-chemistry/biology and legal topics relating to patent information.
October 1, 2010
Biocuration 2010 —The Conference of the International Society for Biocuration
Odaiba, Tokyo, Japan October 11, 2010—October 14, 2010
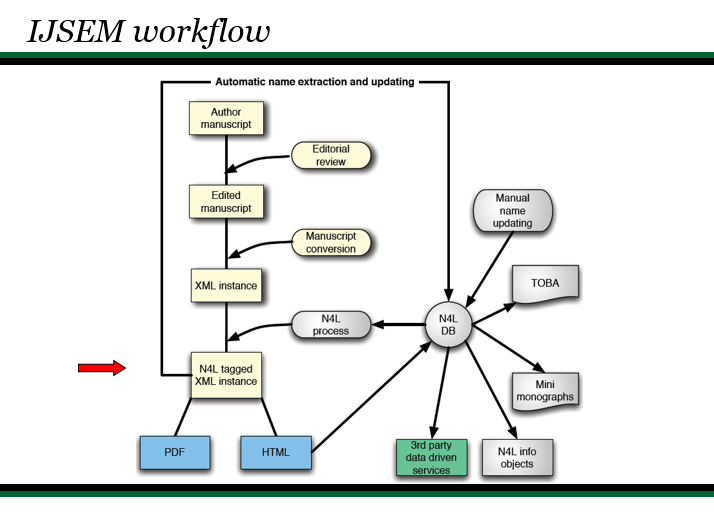
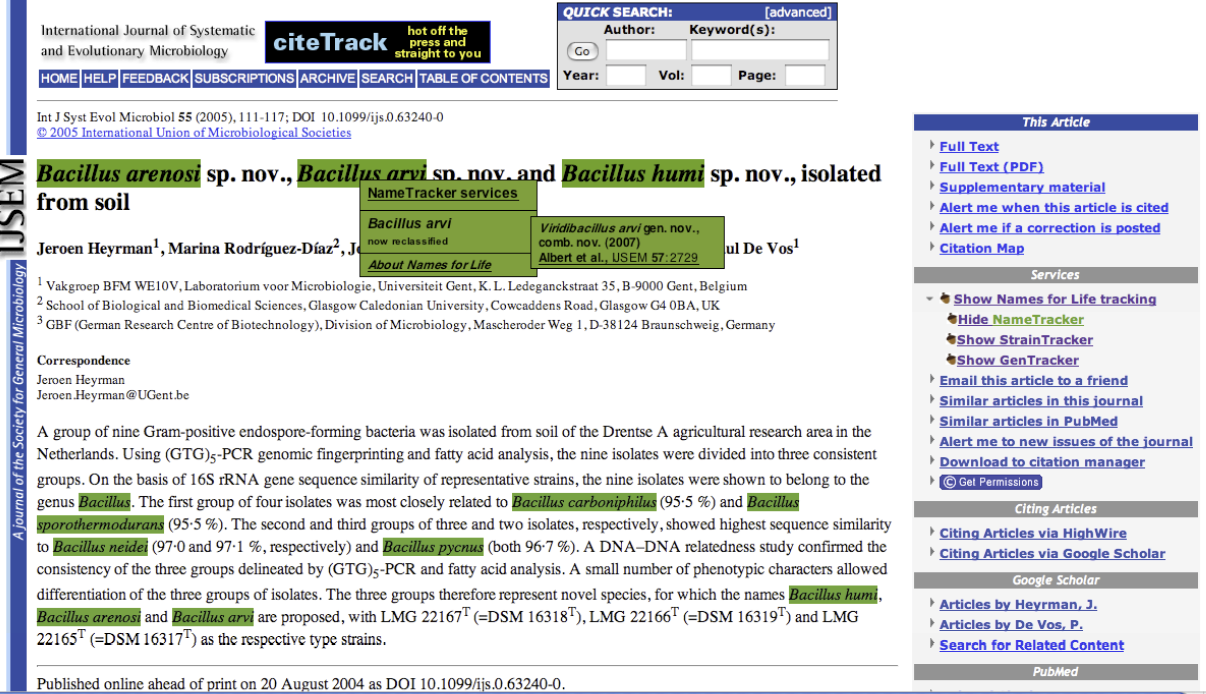
To assist those confronted with ambiguous names (which not only includes researchers but clinicians, manufacturers, patent attorneys, and others who use biological data in their routine work), we developed a generalizable semantic model that represents names, concepts, and exemplars (representations of biological entities) as distinct objects. By identifying each object with a Digital Object Identifier (DOI), it becomes possible to place forward-pointing links in the published literature, in databases, and vector graphics that can be used as part of a mechanism for resolving ambiguities, thereby “future proofing” a nomenclature or terminology. A full implementation of the N4L model for the Bacteria and Archaea was released in April, 2010. The system is professionally curated and represents a Tier III resource in Parkhill’s view of bioinformatic services (Genomic information infrastructure after the deluge, Parkhill et al. 2010). A variety of tools and web services have been developed for readers, publishers, and others (N4L Guide, N4L Autotagger, N4L Semantic Search, N4L Taxonomic Abstracts) and we are incorporating other taxonomies into the N4L data model, as well as adding additional phenotypic, genotypic, and genomic information to the existing exemplars to add greater value to end users.
May 7, 2010
ASM 2010 —American Society for Microbiology 110th General Meeting
San Diego, California May 23, 2010—May 27, 2010
NamesforLife will be attending the ASM 2010 Meeting. Stop by the Society for General Microbiology booth, grab a brochure, sign up for a free account and try live demonstrations of the NamesforLife document annotation and rich content services for publishers.
March 22, 2010
Society for General Microbiology Spring 2010 Meeting
Edinburgh, Scotland, United Kingdom March 29, 2010—April 1, 2010
NamesforLife will have a booth at the SGM Spring 2010 meeting. Please stop by in between sessions to sign up for a free account and try live demonstrations of the NamesforLife document annotation and rich content services for publishers.
February 1, 2010
Genomic Science 2010 —Awardee Workshop VIII and USDA-DOE Knowledgebase Workshop
Crystal City, Virginia February 7, 2010—February 10, 2010
Charles Parker will be presenting poster number 231 (“NamesforLife Semantic Resolution Services for the Life Sciences”, Abstract Book, page 179) in the Tuesday afternoon reception and scientific mixer of the Genomes-to-Life Awardee Workshop.
Please also visit poster 230 (“Standards in Genomic Sciences: Launch of a Standards Compliant Open-Access Journal for the ‘Omics Community”, Abstract Book, page 178) on Monday evening for an update on the recently launched Open Access journal Standards in Genomic Sciences.
Now that the Bacterial Nomenclature database is complete and updated in synchrony with the valid publication of nomenclatural changes, NamesforLife is in the process of linking together Bacterial Nomenclature, technical literature, and the various projects of the Genomes-to-Life program. In N4L, each individual organism is represented by a metadata object (an N4L Exemplar), which is identified by a DOI.
An N4L Exemplar aggregates what is known about an individual organism. The Genomes OnLine Database (GOLD), Standards in Genomic Sciences (SIGS), Genomic Encyclopedia for Bacteria and Archaea (GEBA) and Genomes and Metagenomes Catalogue (GEM) all use unique identifiers that link to each other in some way; via the GCat identifier, GOLD stamp, and GEBA Taxon Identifier. However, there is no single common link to the literature. NamesforLife is closing this gap by tying these disparate sources of information together via N4L Exemplars, which are integrated with the N4L Nomenclature Database and N4L Contextual Index.
The Beta release of the N4L Browser Add-on is officially scheduled to coincide with the Society for General Microbiology conference at the end of March 2010, but it is already available for early testing. Instructions on installation and use can be found at the NamesforLife website. This Firefox Add-on detects and links bacterial names to the N4LDB, providing up-to-date nomenclature, strain and genome information, and a full bibliography.
January 1, 2010
Annual Collaboration for Entrepreneurship 2010
Ann Arbor, Michigan January 20, 2010—January 20, 2010
On Sunday evening, NamesforLife, LLC joined a host of other Michigan-based startup companies exhibiting at ACE’10: The Annual Collaboration for Entrepreneurship in Ann Arbor, Michigan. The event is the culmination of the year-long activities of the Ann Arbor SPARK economic development group, which brings entrepreneurs and investors together in Southeast Michigan for an evening of networking and showcasing.
Charles Parker, the software architect for NamesforLife, reflected on how the Michigan business environment has changed since ACE’09. “A lot of tech companies like Hewlett-Packard have closed sites in Michigan in the past year. The good news is that the tech incubators - SPARK in Ann Arbor, the Technology Innovation Center in East Lansing where we’re located, and others throughout the region, have turned the surplus of local tech talent into an opportunity to invest in home-grown businesses which have a stake in the state economy. Just look around, almost none of the companies here tonight existed a few years ago, and these are all Michigan-based companies.”
May 1, 2009
ASM 2009 —American Society for Microbiology 109th General Meeting
Philadelphia, Pennsylvania May 17, 2009—May 21, 2009
NamesforLife will be attending the ASM 2009 Meeting. Stop by the Society for General Microbiology booth for a live demonstration of the NamesforLife document annotation and rich content services for publishers.
April 2, 2009
United Nations Convention on Biological Diversity —Seventh Meeting: Ad hoc Open-Ended Working Group on Access and Benefit Sharing
Paris, France April 2, 2009—April 8, 2009
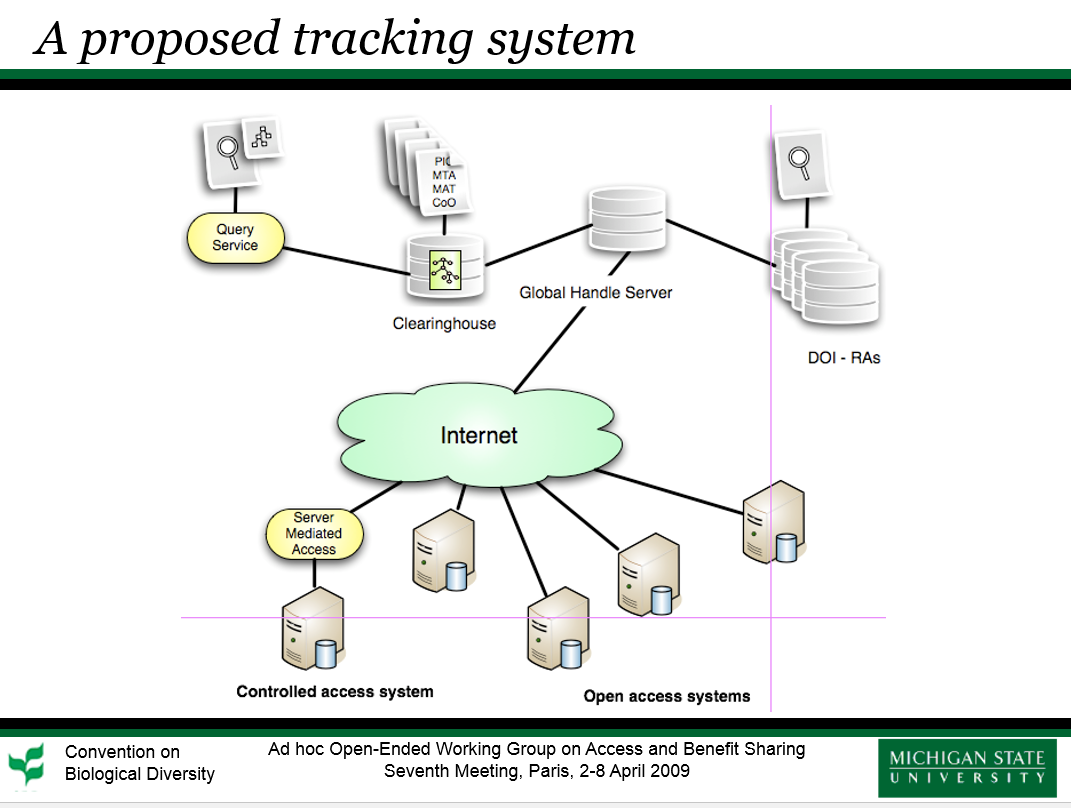
Excerpts from: Studies on the Identification, Tracking and Monitoring of Genetic Resources
After reviewing recent methods of identifying genetic resources directly based on DNA sequences, we have identified methods of tracking and monitoring genetic resources through the use of persistent globally unique identifiers, including practicality, feasibility, costs, and benefits of different options.
Herein, we outline our recommendations for baseline requirements for such a global tracking system to aid users and providers in complying with CBD ABS objectives.
February 8, 2009
Genomics 2009 —GTL Awardee Workshop VII and USDA-DOE Plant Feedstock Genomics for Bioenergy Awardee Workshop
Bethesda, Maryland February 8, 2009—February 11, 2009
Charles Parker will be presenting poster number 135 (“NamesforLife Semantic Resolution Services for the Life Sciences”, Abstract Book, page 182) in the Tuesday afternoon poster session of the Genomes-to-Life Awardee Workshop.
While you’re here please also visit poster 134 (“Release of Taxomatic and Refinement of the SOSCC Algorithm”, Abstract Book, page 180) for updates on the SOSCC algorithm and poster 136 (“Standards in Genomic Sciences: an Open-Access, Standards-Supportive Publication that Rapidly Disseminates Concise Genome and Metagenome Reports in Compliance with MIGS/MIMS Standards”, Abstract Book, page 183) for information on the launch of a new Open Access journal, Standards in Genomic Sciences.
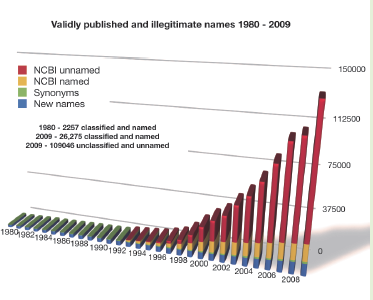
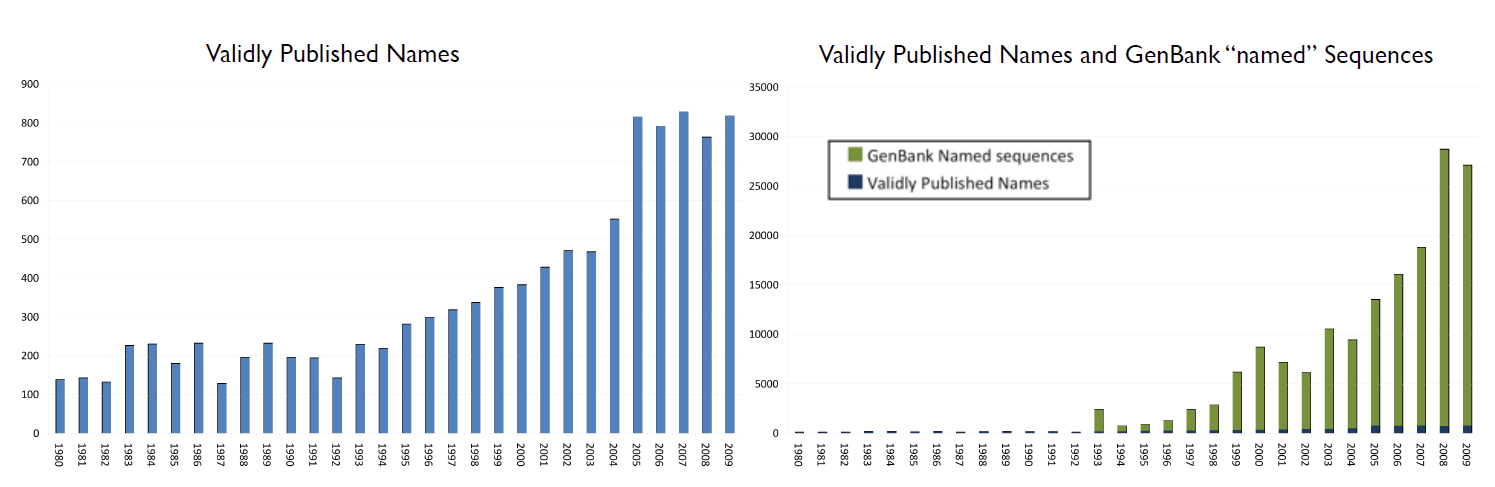
The adoption of DNA sequencing as the preferred method of rapidly characterizing Bacteria and Archaea has tremendously accelerated during the past five years, with the expected consequences. At present, the rate at which “named” sequences are added to the GenBank taxonomy exceeds the rate at which validly published names appear in the taxonomic record by a factor of approximately 35. This confounds the retrieval of related information from various databases and the scientific, technical and medical literature as many of these invalidly named species can not be readily tracked over time, nor can relationships be inferred to those species for which at least one genome sequence is available. This disconnect between the knowledge contained in the literature and the accumulated genomic data is likely to grow as faster and cheaper sequencing methods come into the market place.
The target audience of N4L services is the broad scientific community and others who may need to know the precise meaning of biological names or other terms, in correct temporal context as they are encountered in other digital content (scientific or technical literature, regulatory literature, databases, etc). The dynamic, yet asynchronous nature of biological nomenclature and similar terminology poses a significant burden on information providers, as they must either invest in constantly maintaining their offerings to keep current or shift that burden to their end-users. If the former, the costs can be significant, and, in the absence of a means to synchronize updates across an entire domain of knowledge, end users are still confronted with apparent discrepancies across data sources and content providers. If the burden is shifted to end-users, they must then locate alternative information sources, typically hosted through a web portal, that must be queried separately. This makes utilization of content cumbersome and can lead to considerable ambiguity.
The NamesforLife approach is to semantically enable content in a manner that is transparent to end-users at two points in the value chain: at the source (the data provider or publisher) and at the client side (the end-user). In either case, the end-user experience is the same. At each occurrence of a validly published bacterial or archaeal name, they can have access to precise authoritative information by simply clicking on the name. Tools to enable publishers’ content at the pre-publishing stage that embed persistent N4L identifiers in inline text ensures that their readers will always have access to the correct meaning of the name (as well as additional information), even if the name has changed since publication. Our web-based client supports semantic enablement of other digital content, on-the-fly, providing similar seamless access to NamesforLife content at each point where a validly published name occurs. This provides the reader with direct access to a wealth of information to aid in the interpretation of each enabled article.
January 21, 2009
Annual Collaboration for Entrepreneurship 2009
Ann Arbor, Michigan January 22, 2009—January 22, 2009
On Thursday evening, NamesforLife, LLC joined several other inaugural tenants of newest tech incubator in Michigan (the East Lansing Technology Innovation Center), in attending ACE’09: The Annual Collaboration for Entrepreneurship in Ann Arbor, Michigan. The ACE event, started in January 2001, brings together several Michigan entrepreneurial groups for an evening of networking and showcasing.
May 28, 2008
Society for Scholarly Publishing 2008 Annual Meeting —30th Annual Meeting
Westin Copley, Boston, Massachussetts May 28, 2008—May 30, 2008
George Garrity will be presenting a lecture titled “Say What You Mean: How Semantic Tagging Makes Content More Discoverable, More Useful, and More Valuable” during Seminar 4.
Our next step is to achieve a production-level N4L application (DOI service), which will provide N4L enablement of published STM literature and to investigate other microbiological applications, including a pipeline approach to capture nomenclatural acts and auto-generation of prokaryotic taxonomies. We will also implement a browser plug-in for on-the-fly enablement of web content.
We are actively seeking interested parties to test our tools and concepts.
February 14, 2008
American Association for the Advancement of Science —2007 Annual Meeting
Boston, Massachusetts February 14, 2008—February 18, 2008
George Garrity will be attending the annual meeting of the American Association for the Advancement of Science.
February 10, 2008
Genomics 2008 —GTL Awardee Workshop VI and Metabolic Engineering Working Group Interagency Conference on Metabolic Engineering
Bethesda, Maryland February 10, 2008—February 13, 2008
George Garrity will be presenting poster 142 (“NamesforLife Semantic Resolution Services for the Life Sciences”, Abstract Book, page 136) at the Tuesday evening poster session (5:00pm-8:00pm) in Salon ABCD.
While you are here, please also visit poster 141 (“Further Refinement and Deployment of the SOSCC Algorithm as a Web Service for Automated Classification and Identification of Bacteria and Archaea”, Abstract Book, page 135) during the Monday evening poster session (5:00pm-8:00pm), also in Salon ABCD.
Within the Genomes-to-Life Roadmap, the DOE states that a significant barrier to effective communication in the life sciences is a lack of standardized semantics that accurately describe data objects and persistently express knowledge change over time. As research methods and biological concepts evolve, certainty about correct interpretation of prior data and published results decreases because both become overloaded with synonymous and polysemous terms. Ambiguity in rapidly evolving terminology is a common and chronic problem in science and technology. NamesforLife (N4L) is a novel technology designed to solve this problem.
April 21, 2007
Mid-Michigan Entrepreneur's Day
East Lansing, Michigan April 25, 2007—April 25, 2007
George Garrity presents the NamesforLife business model at the Mid-Michigan Entrepreneur’s Day.
NamesforLife, LLC is initially pursuing commercialization in the Scientific, Technical and Medical (STM) publishing sectors, as well as Biological Resource Centers (BRCs) and diagnostic equipment vendors.
In the longer term, we are looking to adapt the NamesforLife model to other terminologies and nomenclatures for economically important eukaryotes, genome annotation and medical/pharmaceutical terminology.
February 7, 2007
Food and Agriculture Organization (FOA) of the United Nations —IT Support for SMTA implementation
Rome, Italy February 14, 2007—February 14, 2007
George Garrity provides some thoughts on the application of persistent identifiers to Standard Material Transfer Agreements (SMTAs).
NamesforLife provides a method for persistently linking the occurrence of a biological name or other technical term in third party content to managed information about its origins, formal definition, current usage, and related goods and services. This Information Architecture is based on some of the properties of persistent identifiers, and our implementation specifically uses Digital Object Identifiers to link hetereogeneous data and resolve ambiguous names.
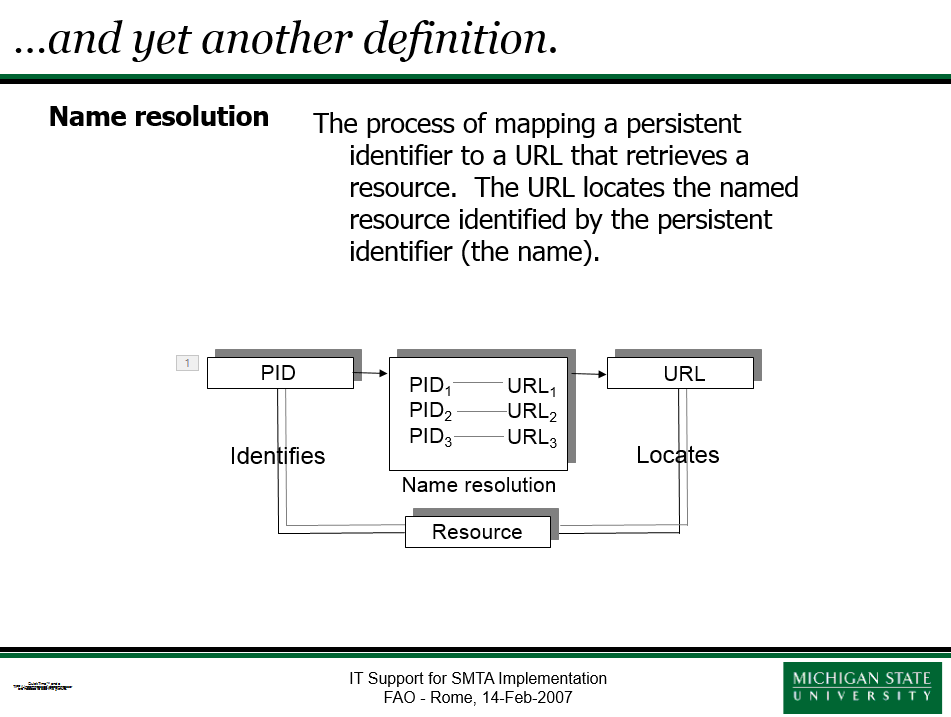
January 5, 2007
The ABS Dialogues —The Role of Documentation in ABS and TK Governance
Hotel Plaza del Bosque, Lima, Peru January 21, 2007—January 21, 2007
George Garrity presents the lecture “An Overview of Persistent Identifiers” in the afternoon meeting, “New approaches to documentation of genetic resources”.
A persistent identifier (PID) has one or more of the following properties:
- Semantically Opaque (the identifier avoids any embedded meaning)
- Governance (a technical and/or social framework oversees development, implementation and “marketing” of the identifier)
- Persistence (a mechanism guarantees persistence of issued identifiers)
- Registration (a mechanism exists for global registration of identifiers)
- Metadata (minimal requirements exist for metadata associated with each identified object)
- Standardization (the identifier conforms to an accepted standard)
- Globally Unique (the identifier is globally unique)
- Widespread Usage (the identifier is in widespread usage)
- Object/Location Resolution (the identifier actually identifies something)
- Actionable (network services are attached to the identifier)
- Uniqueness (a resolution service checks for uniqueness at the local level)
- Interoperability (the identifiers are readily incorporated into other applications without modification or permission)
- Granularity (the identifiers can be assigned to subcomponents (nesting of entities within entities))
- Business Model (a compelling business need ensures that the identifier infrastructure can be maintained in a self-supporting manner)
The Digital Object Identifier (DOI) exhibits all of these characteristics.

September 7, 2006
eGenomics 2006 —eGenomics III: Cataloguing our complete genome collection
Robinson College, Cambridge, United Kingdom September 11, 2006—September 13, 2006
George Garrity discusses NamesforLife and PhenBank at Cambridge. He will also chair Monday’s second session: “Databases and Metadata capture and Exchange efforts”.
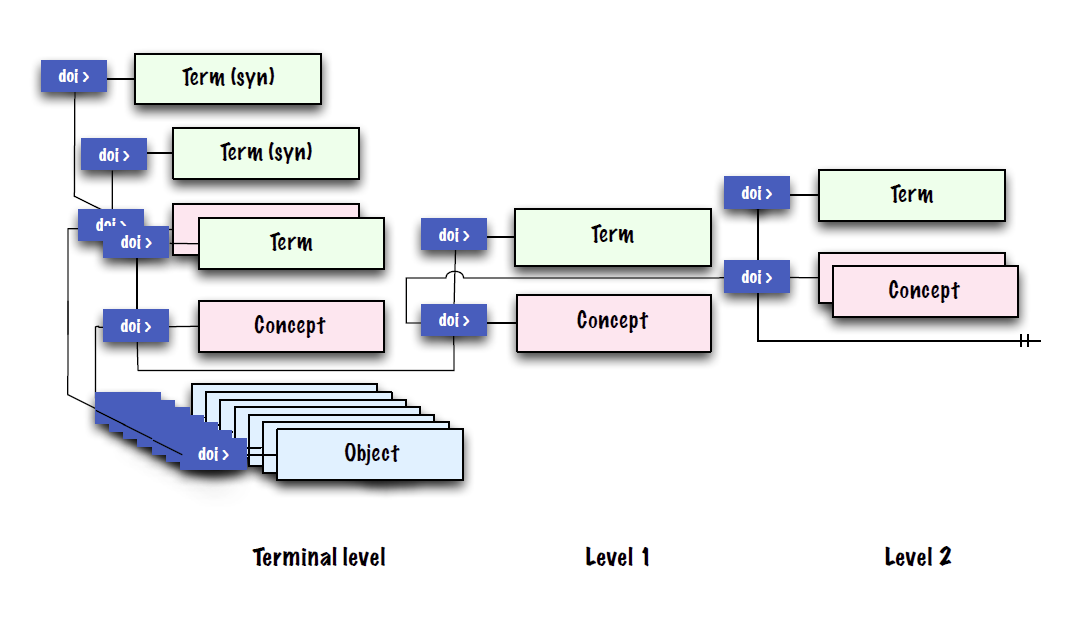
Names, taxon concepts and exemplars are independent. Names are fixed in time and are bibliographic events, tied to a particular published description. The taxon concept, however, drifts once it comes into usage, as non-type exemplars are added to the global sample set. There is also a critical need to always tie the data (phenotype and genotype) to the correct source strain.
When one looks at the environmental data, it becomes difficult to accurately interpret results across studies, especially when one is dealing with survey data comprised of a single measurement (e.g., a 16S rRNA sequence). One of the reasons is that investigators use their own identifier to label the data (and strains). More importantly, many of these labels are not unique.
We are in the process of updating our prototype to identify all of the high quality 16S rRNA sequences that have come from type strains held in different Biological Resource Collections (BRCs).
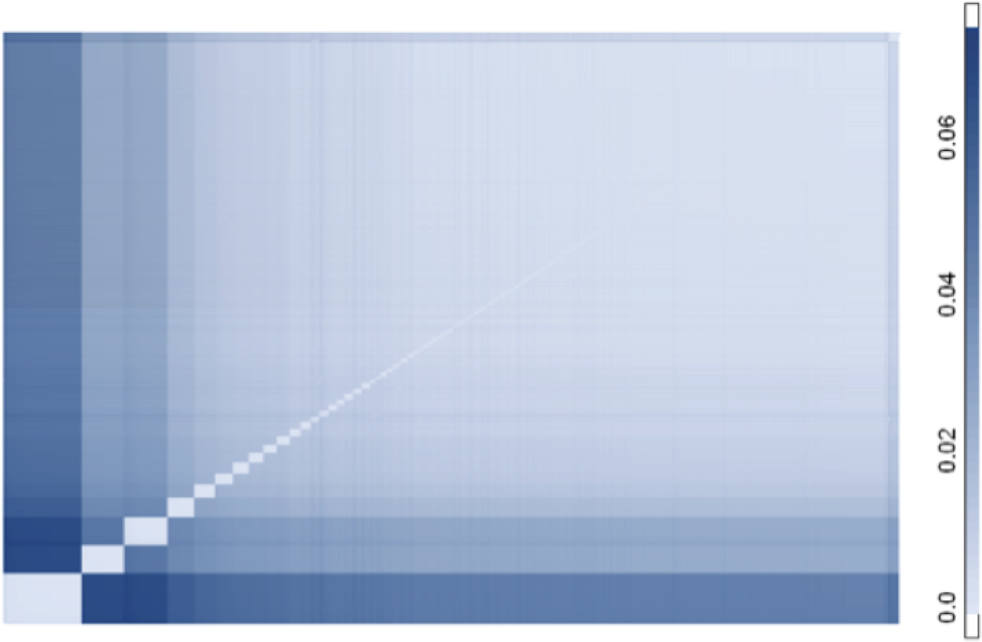
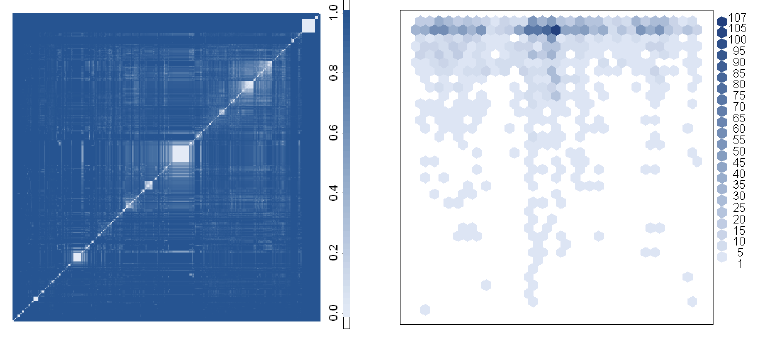
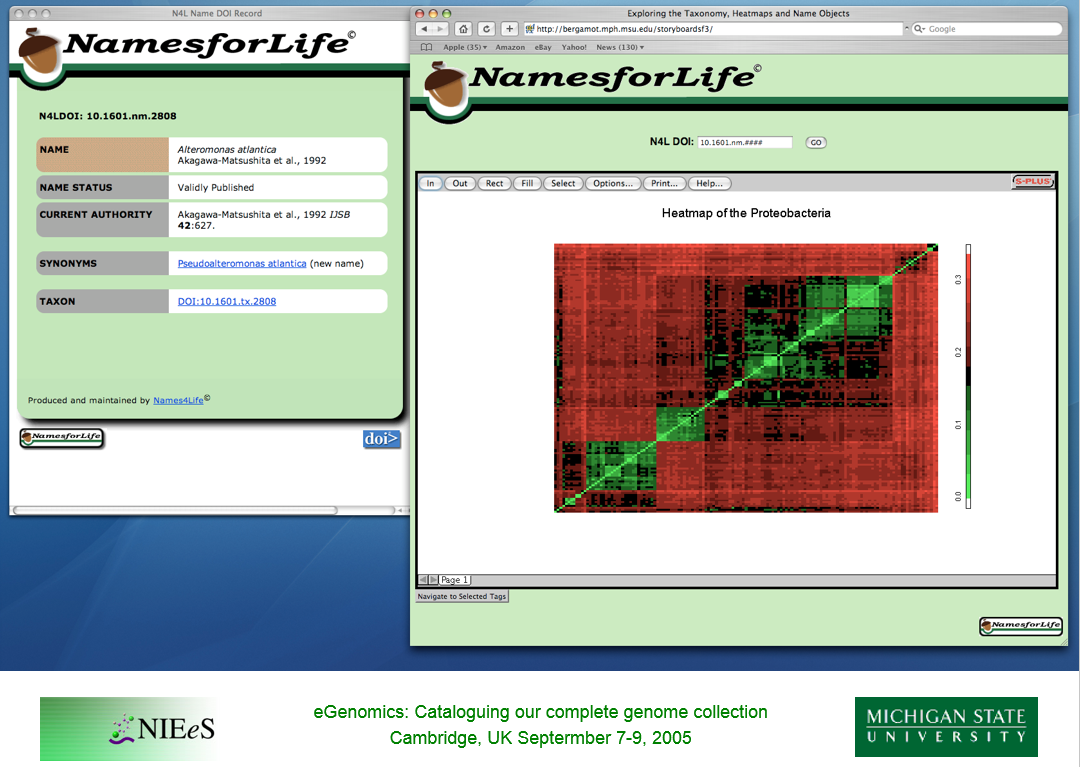
We have been using heatmaps of evolutionary distance matrices to visualize sequence similarity and to uncover annotation errors in the 16S rRNA sequence data set for about five years. Last year, we published the SOSCC algorithm which can undertake this process in an automated manner.
What is particularly useful is that the method allows us to examine 1,000–10,000 sequences simultaneously, thereby revealing the otherwise hidden structure associated with more distant taxonomic relationships.
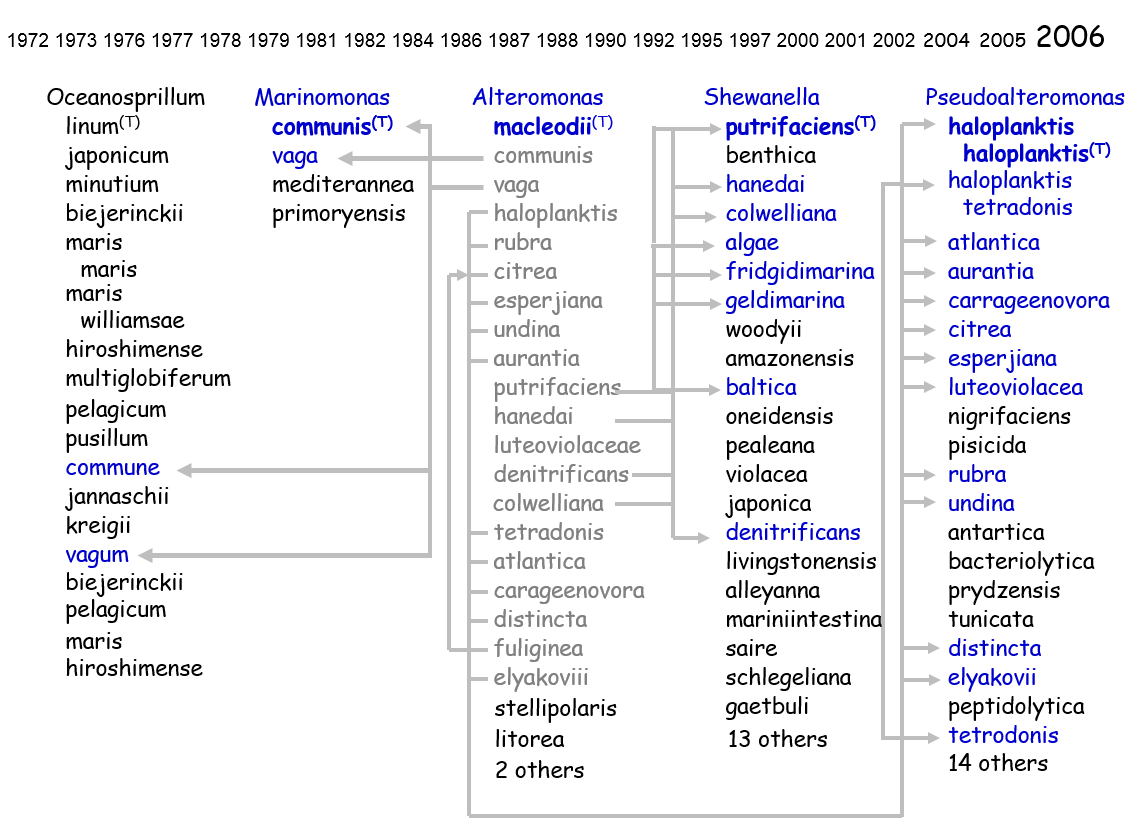
July 8, 2006
2nd FEMS Congress of European Microbiologists —Integrating Microbial Knowledge into Human Life
Madrid, Spain July 4, 2006—July 8, 2006
George Garrity presents “Knowledge bleed, PhenBank, and NamesforLife” during Symposium 20 (Biodiversity).
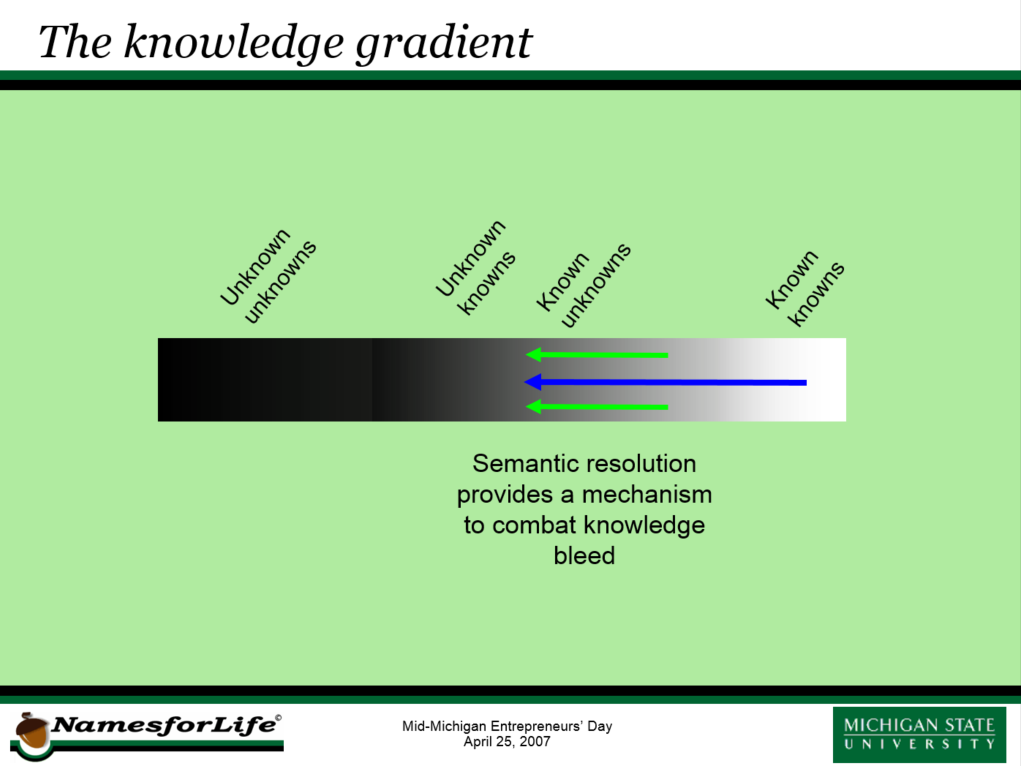
There are different scopes of knowledge. There are those things that we know that we clearly understand. There are also those things that are totally unknown to us. Research helps to increase our fundamental knowledge, pushing back the boundaries of our ignorance and creating a third category of knowledge, those things that we do not yet know, but which we know we do not know them.
It is our opinion that within the knowledge gradient, there exists another type of unknown - representing knowledge that was once known, but has been forgotten or lost over time. We call this the “unknown knowns”. At first glance, this might seem an implausible, but it represents a very real risk, not only in biodiversity studies, but in most fields, with the biosciences being the among the most prone to this problem, because of the extraordinary growth in many of the sub-disciplines, and the accompanying way of reporting results. A principle source of this knowledge loss arises in the very terminology we use to discuss and report our findings. Unless each worker clearly understands the underlying concepts that are used to describe their work in reference to that of others, discovery and retrieval of important findings becomes more difficult, if not impossible. Part of the problem lies in the sheer volume of material that is appearing in “print”. The second involves the rapidly evolving terms that are used to describe biologically relevant concepts at the various levels.
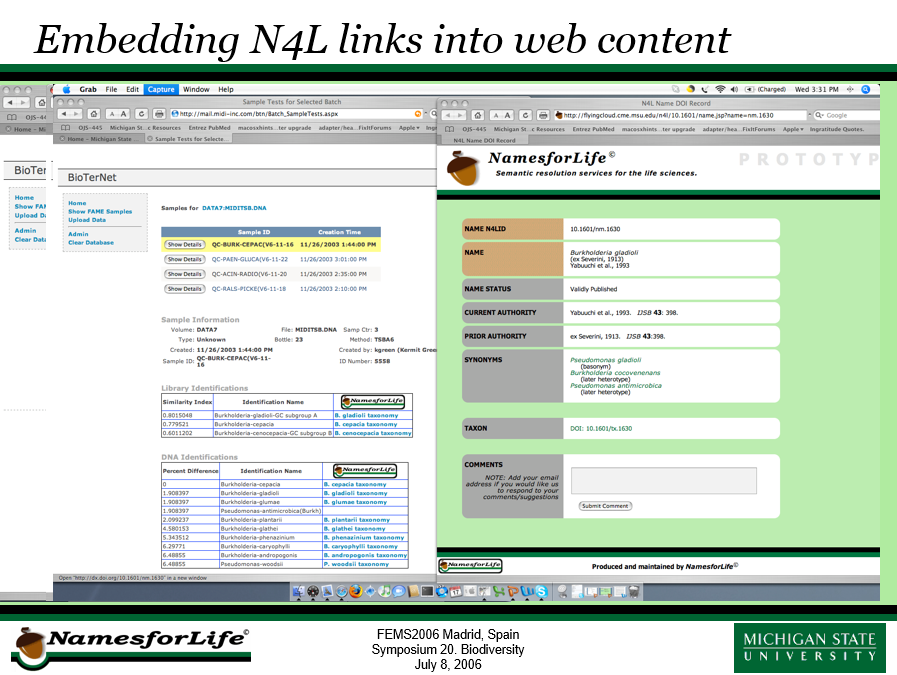
February 1, 2006
Taxonomic Databases Working Group GUID-1 Workshop —First International Workshop on Globally Unique Identifiers (GUIDs) for Biodiversity
National Evolutionary Synthesis Center (NESCent), Durham, North Carolina February 1, 2006—February 3, 2006
George Garrity presents unveils a working prototype of the NamesforLife Information Architecture.
In January, we launched a working prototype of an Information Architecture (IA) based on the NamesforLife (N4L) Model. This architecture provides a transparent information layer to deliver Digital Object Identifier (DOI) services to the life science community. The architecture also implements an ontology with a schema that produces metadata consistent with requirements of the International DOI Foundation (IDF). The initial services will conform to DOI Application Profile (AP) 0.
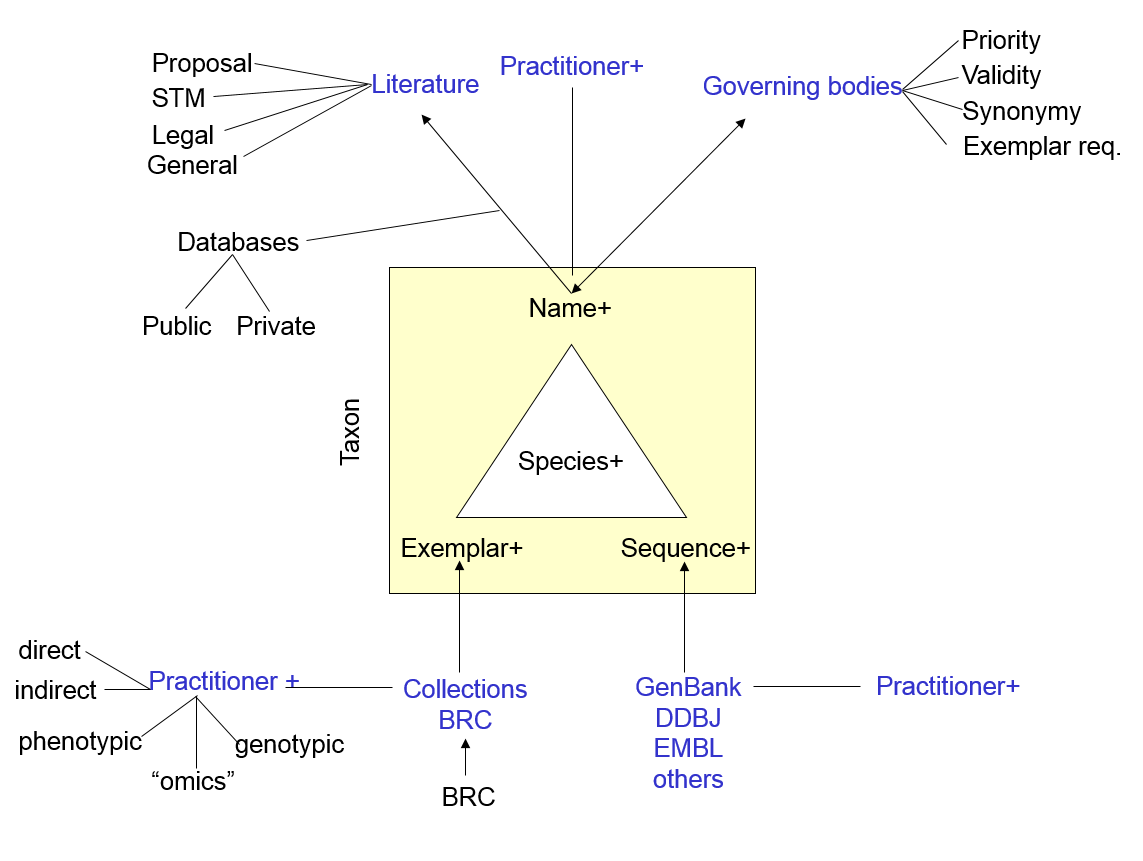
This test case contains 24,176 first-class objects comprising: Name, Taxon, Exemplar, Nomos, Practitioner, Feature, and Nomenclatural Code. This system is based on a nomenclatural taxonomy, but capable of supporting multiple taxonomic views and “time travel”, which will enable us to track changes in concepts over time.
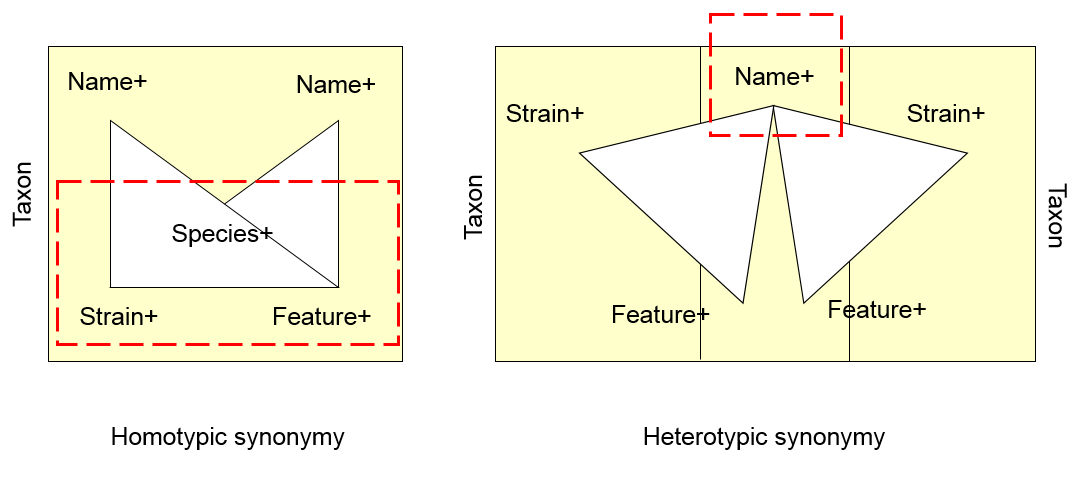
September 7, 2005
eGenomics 2005 —eGenomics II: Cataloguing our complete genome collection
Centre for Mathematical Sciences, Cambridge, United Kingdom September 7, 2005—September 9, 2005
George Garrity describes progress on the NamesforLife proof-of-concept and proposes the idea of PhenBank, a phenotypic data repository, at Cambridge.
The currently available taxonomic data sources have an unlimited number of data types, some of which are broadly applicable across all taxa, most of which are not. Some are cumulative, many are comparative. There exist numerous taxon-specific vocabularies, and there are few links to primary literature or original data sets. Existing tools for working with phenotypic data are of variable quality, most are “one-off” and non-interoperable. Fixing these problems has limited public support, since the user bases and data curation varies with economic importance, thus funding is poor to non-existant.
We propose a public repository for phenotypic and taxonomic data that adheres to a common data model and provides a source of interoperable phenotypic data for the Microbiology community.
July 1, 2005
International workshop (IUAP V/23) —Exploring and exploiting microbiological commons: contributions of bio-informatics and intellectual property rights in sharing biological information
University Foundation, Egmontstraat 5, Brussels, Belgium July 7, 2005—July 8, 2005
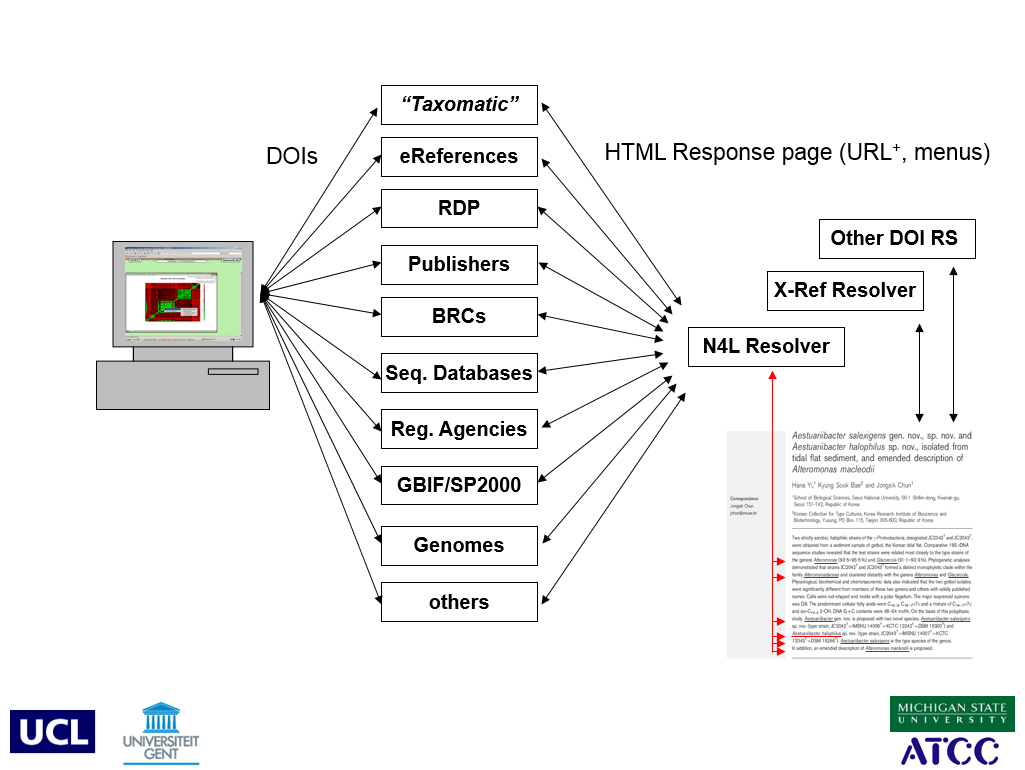
George Garrity presents the N4L system in “Automating the Quest for Novel Prokaryotic Diversity (revisited)”.
Previously, we demonstrated the value of using techniques drawn from the field of Exploratory Data Analysis (EDA) for the analysis and visualization of large sets of sequence data (notably SSU rRNA gene sequences) that are used to construct a comprehensive taxonomy of prokaryotes. While the approach is computationally efficient and quite useful in uncovering a variety of taxonomic and annotation errors, the methods suffered from some practical limitations; notably bottlenecks in the preprocessing of data for our analyses. Work is currently underway to address these limitations that will greatly expedite the preprocessing steps through a pipeline approach. In addition, new methods are under active development that will automatically flag misidentified and potentially novel sequences within a given dataset and automatically place such sequences into close proximity to their nearest neighbors, based on 16S rDNA sequence homology. These methods will also permit linking of EDA plots, derived from such analyses to external data and information resources.
March 14, 2005
Bioinformatics Forum —Names and Objects for Unambiguous Data Access amongst Biodiversity Data Entities
National Institute for Environmental Studies, Tsukuba, Ibaraki, Japan March 14, 2005—March 15, 2005
Catherine Lyons presents “An Introduction to Digital Object Identifiers as background to NamesforLife”.
Systematic taxonomy is a complex network of documents, data, and, concepts. The Digital Object Identifier (DOI) system is built from components that model complexity in other domains. This is an unusual introduction to DOIs, in that it emphasizes those aspects of the DOI system that will be a particular strength in the management of taxonomy and nomenclature. The association of objects with types, and types with type-specific metadata, enable a DOI ‘Application Profile’ (AP). An AP gathers together digital objects that have common metadata properties. For a DOI in a given AP, a service can be implemented that exploits the metadata defined by its AP, and returns, for example, some text, a link, a menu.
Suppose there were a Biological Name AP associated with a ‘Check for Synonyms’ service...this service could be associated with digital objects (Information Objects) in the Name AP (i.e., nomenclatural assertions). By reasoning over Information Objects, we can construct services that can be offered through multiple resolution.

June 22, 2004
Annual International DOI Foundation Members Meeting —Session 4: Uses of identifiers - Identifiers for data
London, United Kingdom June 22, 2004—June 22, 2004
Catherine Lyons presents the NamesforLife concept at the IDF Members Meeting.
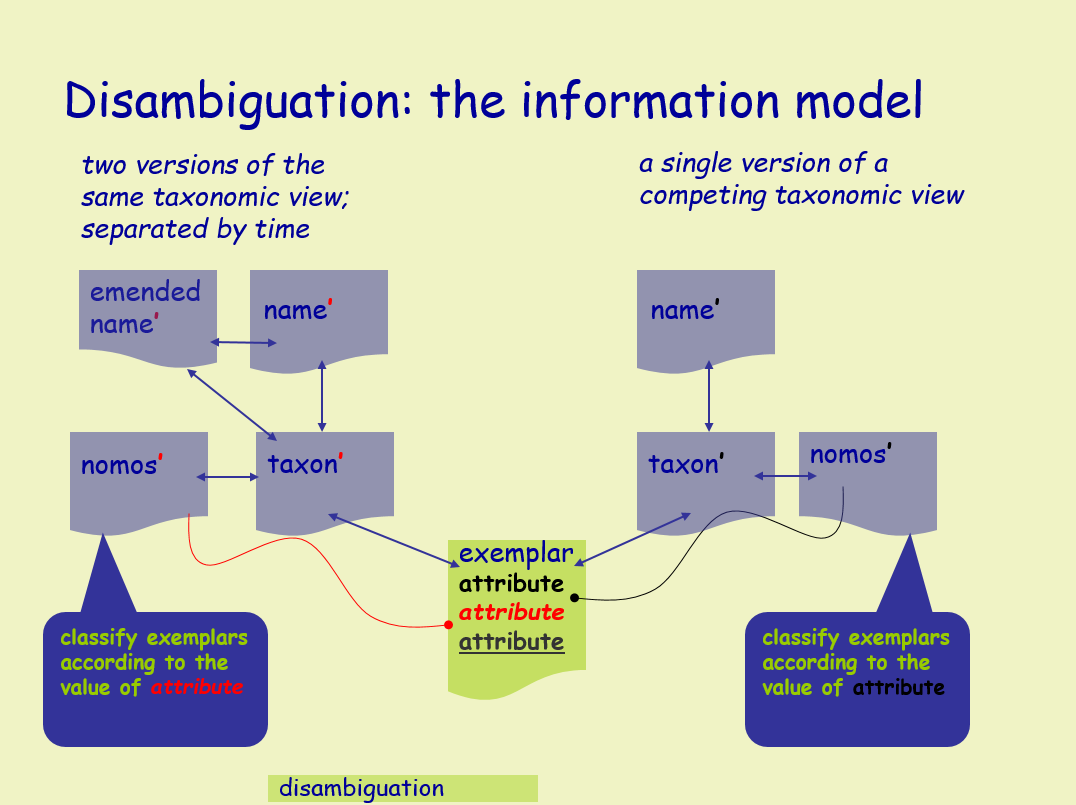
The Alteromonadales represent an interesting test case for demonstrating how one could apply Digital Object Identifiers (DOIs) to solve the problems associated with changes in nomenclature and taxonomy of a particular group. The family was effectively defined by Garrity et al. in version 1.0 of the Taxonomic Outline and independently by Ivanova and Mikhailov in 2001 and is formed on the genus Alteromonas, which serves as the type genus for the family and class. Alteromonas was initially circumscribed by Bauman et al. in 1972 and subsequently emended (although not formally in all cases) on more than 15 occasions through the addition 20 species. Nineteen of these species were subsequently moved to four other genera, two of which are also members of the Alteromonas (sensu Garrity et al.) and two genera are members of the family “Oceanospirillacea”, class “Oceanospirillales”. Some of the later proposals also yield three heterotypic synonyms, two homotypic synonyms, the subdivision of one species into two subspecies which were subsequently rejoined following a move to another genus, the subsequent subdivision of one reassigned species into five distinct species in that genus, and one orthographic correction that was required to correct an error when latinizing a species name. Thus, the original 20 species of Alteromonas have appeared under a total of 64 different names in five genera, two families and two classes.
If we apply an Information Model based on the separation of the Names (labels), Taxa (concepts), and Exemplars (strains/objects), we are able to track changes in nomenclature and taxonomic opinion separately, without losing track of the underlying organism (the Exemplar). This enables a means of separating competing taxonomic views, thereby effectively disambiguating any synonymous names and competing taxonomies applied to an exemplar.
Further, if we assign a DOI to each Name, Taxon, and Exemplar, we essentially create a set of Information Objects - persistent, online, public documents - which serve to instantiate nomenclatural events, taxonomic opinions, and exemplars. These Information Objects provide metadata and form a navigable graph when linked with other Information Objects and to online information outside of NamesforLife. They are easy to link to from online journals, databases, and similar resources, and are guaranteed to be persistent.
To achieve a working prototype based on this Information Architecture, we plan to perform some exploratory work with publishers, biodata curators and genomics researchers to find a path toward obtaining funding for this project and developing standards for clean nomenclatural and taxonomic data.
May 23, 2004
ASM 2004 —American Society for Microbiology 104th General Meeting
New Orleans, Louisiana May 23, 2004—May 27, 2004
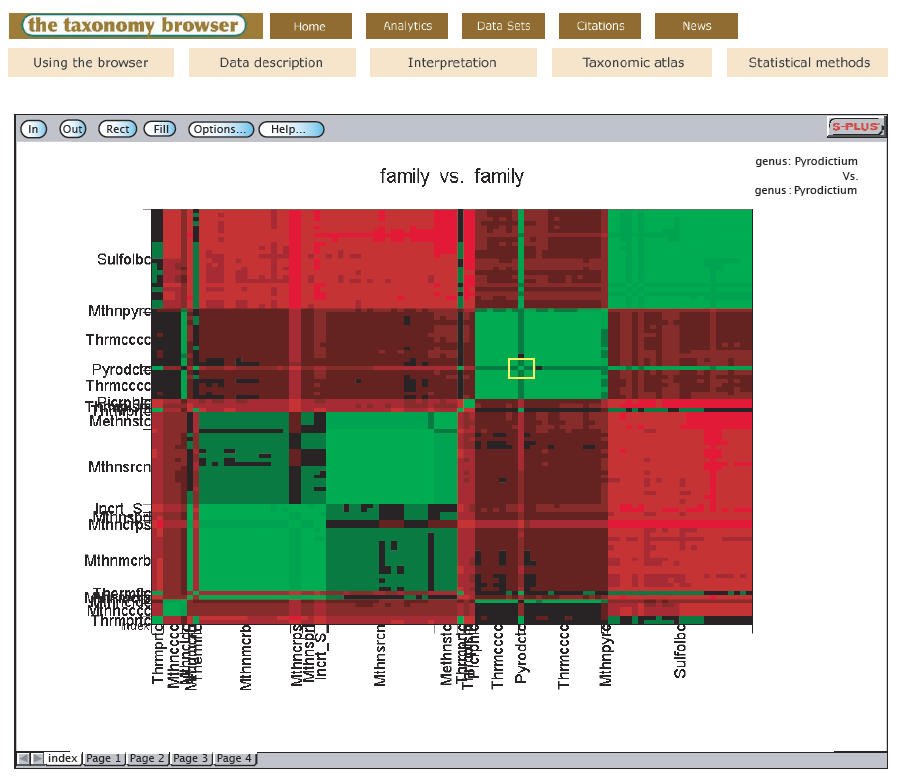
Dr. Garrity will be presenting a taxonomy browser backed by a novel algorithm for building self-organizing and self-correcting classifications.
Recently, we developed an algorithm that builds self-organizing and self-correcting classifications. We have applied this algorithm to the problems arising from sequence annotation errors on prokaryotic classification. The comparison of the optimized classifications developed with our algorithm with other taxonomic proposals has allowed us to resolve outstanding problems in prokaryotic classification and taxonomy.
To make such comparisons available to the research community, we have built a website that allows users to compare the current Bergey’s Taxonomic Outline with an optimized classification. The website serves as user interface to a dedicated analytic server, built using StatServer (Insightful). The application allows users to select the taxonomic group they are interested in, choose how they want the results to be organized (that is, at the species, genus or family level) and display the comparison. The organization of the compared classifications is visualized in the form of shaded evolutionary distance matrices. The colors of the matrix indicate the distances between the pairs of sequences in the matrix. The grouping of the colors in the matrix reflects the higher level groupings of the sequences (and, by extension, of the parent organisms). One matrix is arranged according to the hierarchy of the Outline and the other matrix is arranged according to the groupings generated by the classifier. Users can drill down in the display to see the comparisons at lower taxonomic levels or move up the hierarchy. The side-by-side comparison illuminates possible solutions to evident problems in the current classification. We illustrate how the taxonomy browser works by looking at the classification and taxonomy of the Archaea.
October 27, 2003
GBIF/WFCC/SPO Expert Workshop —Towards a Global Infrastructure for Microbial Information
Hotel Metropole, Brussels, Belgium October 27, 2003—October 28, 2003
George Garrity presents “Biological nomenclature in the postgenomic era: Biological and computational issues”.
Within biology, the fundamental taxonomic unit is the species. However, species can be further subdivided into subspecies, varieties and other categories that are specific to the disciplines of botany, zoology, prokaryotic biology and virology. In the preferred example, the species are within the domains Bacteria and Archaea, which are collectively referred to as prokaryotes.
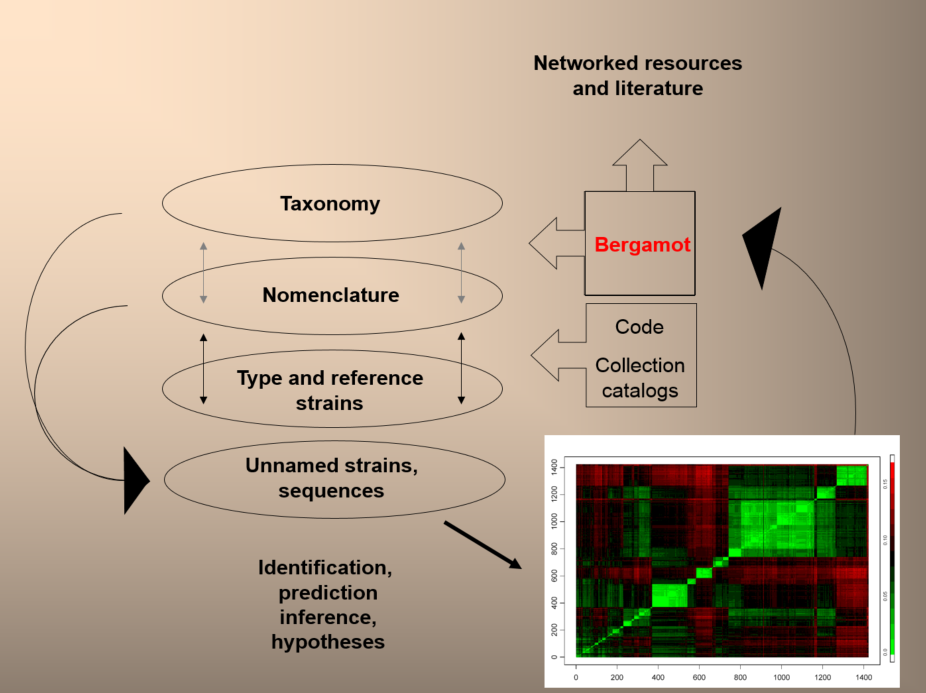
The N4L/Bergamot model and Information Objects provide a transparent middle layer that permanently links together Names and Taxa (at all levels of the hierarchy) with their occurrences in the literature and data repositories. Through the use of DOIs and multiple resolution technology, Names can serve as future-proof links to the complete taxonomic record of a given taxon (including relevant information regarding synonymies, orthographic errors, priority, etc.) and to a variety of third-party services specific to a given taxon without the intervention of search engines or other methods. End-users simply need to click on a name or other similar graphic device to gain access to the desired information.
February 9, 2003
Genomes to Life Contractor-Grantee Workshop I —Workshop Breakout Session - Comparative Genomics: New Approaches & Insights
Arlington, Virginia February 9, 2003—February 12, 2003
George Garrity presents “Carolus Linnaeus in the postgenomic era”.
This discussion will focus on a problem that plagues us all to some degree or another - biological nomenclature. Ideally, our formalized system of nomenclature is supposed to improve communication among biologists. In reality, it seems to be a major obstacle, especially when misapplied. Although the problem is evident in the literature, it is most severe in the sequence databases, which now serve as the principal source and repository of data used in comparative biology. Moreover, the sequence databases tend to propagate such errors for a variety of reasons. As biological data proliferates and interconnects, it depends increasingly on software infrastructure, and it becomes increasingly obvious that biological names do not meet the requirements of a good identifier, in strict computing terms. A good identifier should be unique and persistent. As an outgrowth of my current DOE funded project, we have been exploring a practical and workable solution that we believe will help solve the problem in a future-proof fashion.
February 2, 2003
Workshop on Data Management for Molecular and Cell Biology
Lister Hill Center, NLM, NIH Campus, Bethesda, Maryland February 2, 2003—February 3, 2003
George Garrity will be present to discuss the white paper, “Future-proofing biological nomenclature”.
The disjunction of nomenclature and taxonomy results in an accumulation of names of dubious value in the literature and databases. While systematic biologists may be adept at recognizing such problems, most others (including the curators of some databases) are not.
It is becoming increasingly obvious that biological names do not meet the requirements of a good identifier, in strict computing terms. A good identifier should be unique and persistent. As new data become available, the inferred relationships among the named entities may change: a taxon may be promoted or demoted, new taxa may be interposed between formerly contiguous taxa. As a result, the association of names with taxonomic concepts tends to weaken as the rate at which gene sequencing accelerates. Failure to address this problem will result in increasingly unpredictable responses when biological names are used to query either the literature or databases. What is required is a resolution system that can handle the complex relationships between biological names and the entities they denote and provide links to both the historical and current definition of each named taxon.
We believe that an implementation of the Digital Object Identifier (DOI) may provide the most robust and future-proof solution to this problem. A DOI is a unique, persistent identifier of an information resource that is registered together with a URL. Its purpose is the management and retrieval of that resource in the networked environment. In practice, most current DOIs identify journal articles, but DOIs are now being applied to trade publications, stock photography, and physicochemical data sets.